Efficient Data Management in R
The software environment R is widely used for data analysis and data visualization in the social sciences and beyond. Additionally, it is becoming increasingly popular as a tool for data and file management. Focusing on these latter aspects, this Methods Bites Tutorial by Marcel Neunhoeffer, Oliver Rittmann and our team members Denis Cohen and Cosima Meyer illustrates the workflow and best practices for efficient data management in R.
Readers will learn about:
- the workflow for organizing and conducting complex analyses in R
- creating, editing, and accessing directory hierarchies and their contents
- data merging, data management and data manipulation using tidy R and base R
- the basics of programming and debugging
To illustrate these steps, we will work through an example from comparative political behavior – the question under which conditions center-left parties can successfully mobilize votes among the unemployed. To tackle this question, we will combine micro-level voting data from Round 9 of the European Social Survey with contextual data on election dates from ParlGov, party positions from the Manifesto Project, and unemployment rates from the World Bank.
Note: This blog post is based on our workshop in the MZES Social Science Data Lab in Spring 2020. The corresponding workshop materials can be found on our GitHub.
Contents
Setup
Over the course of this tutorial, we will use the following R packages:
Code: Packages used in this tutorial
## Save package names as a vector of strings
pkgs <-
c(
"foreign", ### read data stored in various formats
"readstata13", ### read data stored by Stata 13-16
"reshape2", ### flexibly reshape data
"countrycode", ### convert country names and country codes
"lubridate", ### dates and time
"dplyr", ### tools for data manipulation
"magrittr", ### piping operations
"tidyr", ### tool to deal with messy data (and get "tidy data")
"ggplot2", ### data visualization using a grammar of graphics
"fabricatr" ### imagine your data before you collect it
)
## Install uninstalled packages
lapply(pkgs[!(pkgs %in% installed.packages())], install.packages)
## Load all packages to library and adjust options
lapply(pkgs, library, character.only = TRUE)Workflow for a Reproducible Data Project
Even the setup of a reproducible data project should follow reproducible steps. And since we need to repeat these steps every time we start a new data project, it is a good idea to keep everything simple and to automate what can be automated. For the following steps to work you need three things:
- A github.com account.
- A current installation of R and RStudio on the computer you want to work on.
- An installation of git on the computer you want to work on.
If you struggle with any of those three prerequisites, happygitwithr.com is an excellent resource that includes a step-by-step guide of how to setup git with R. It will also give you are more in-depth introduction to what git and github actually are and why it is worthwhile including them in any data project right from the start!
Git is a version control system that makes it easy to track changes and work on code collaboratively. GitHub is a hosting service for git. You can think of it like a public Dropbox for code. As a student, you even get unlimited private repositories which you can use if you don’t feel like sharing your code with the rest of the world (yet).
Let’s start a new project:
- Start a new project on GitHub:
- Login to your GitHub account.
- Click on
Newon the left side where your repositories are displayed. - Give your project repository a meaningful name. For this course you could name it:
efficient-data-management. - Click
Create repository. - Copy the URL to the repository, it will look something like this:
https://github.com/<username>/efficient-data-management.git
- Open the project in RStudio:
- Open RStudio.
- Click on
File > New Project.... - Create a new project from
Version Control. - Choose
Git. - Paste the URL to the repository in
Repository URL. - Choose a folder on your computer where the project will be stored locally.
- And finally click on
Create.
Initializing the project with a .Rprofile.
Now you have a new Rproject that is linked with GitHub. Before start coding, we want to have an efficient folder structure. Ideally, we want to use the same structure for all of our projects. This way, it is easier to focus on the things that really matter – like producing interesting research. To that end, we initialize our Rproject with a .Rprofile that automates the process of setting up our project structure.
- Initialize your Rproject with a .Rprofile:
- If you follow these steps for the very first time, install the
renvpackage in R. - In RStudio, with your new Rproject open, open a new
Text File. - Paste the following code and save it as
.Rprofile. - Now close RStudio and re-open your project from your project folder.
- If you follow these steps for the very first time, install the
.First <- function() {
dir.create(paste0(getwd(), "/figures"), showWarnings = F)
dir.create(paste0(getwd(), "/processed-data"), showWarnings = F)
dir.create(paste0(getwd(), "/raw-data"), showWarnings = F)
dir.create(paste0(getwd(), "/scripts"), showWarnings = F)
dir.create(paste0(getwd(), "/manuscript"), showWarnings = F)
if (!("renv" %in% list.files())) {
renv::init()
} else {
source("renv/activate.R")
}
cat("\nWelcome to your R-Project:", basename(getwd()), "\n")
}R will automatically source the .Rprofile at startup. Here, we first create some folders with dir.create(). Don’t worry, folders will not be created if they already exist, so this will not overwrite existing files. We then display a nice welcome message with cat() and activate the renv package.
After re-opening the Rproject, you should now see the project setup in your folder.
What is renv?
In the past, sharing your project with someone else and getting R to show the exact same behavior on different machines could be a pain. The renv package makes this super easy. It creates a local package directory for your project. This means that it keeps track of all the packages and package versions that you use in your project. If someone else wants to work with the exact same package environment to reproduce your data project, they can easily restore it from your renv package directory. To learn more about renv, check out its documentation here.
Leveraging git and renv at the end of a working session
Now that we have our nice project setup, we should not forget to leverage it. At the end of a working session you should follow the following steps:
- Create a
renv::snapshot()to save all the packages you used to your local package directory. - Commit all your changes to git. This can be easily done by using the
Gitpane in RStudio. - Push everything to GitHub.
Whenever you re-open your Rproject, make sure to start your working session with a Pull from GitHub. That way, you will always work with the most recent version of your project.
Managing and Accessing Directories
Now that we have a directory structure for our project, we need to learn how to work with it. What goes into the different folders should be rather self-explanatory. Yet, it is good practice to also include a simple ReadMe file in each of the folders to explain what it should contain.
What is a working directory?
Your working directory is the folder on your computer that R will use to read and save files. With your .Rproject and .Rmd files, you do not have to worry about this too much. There is only one golden rule: Never change your working directory (especially, never to absolute paths specific to your computer).
Where is your working directory?
When we created our .Rproject we had to decide where all the files should be stored. If you open your .Rproject, the working directory will automatically be set to this folder. If you have a .Rmd file somewhere in your project (e.g. in the /manuscript folder) RStudio will automatically set the working directory the folder containing the .Rmd.
What does this all mean?
This means you have to access all data or code (stored in the folders of your project) relative to your current working directory.
- If you work with a simple
.Rscript (and you store the file in/scripts) in your.Rprojectand you want to open data stored in your/raw-datafolder: Simply open the file directly from the folder e.g. withread.csv("raw-data/20200127_parlgov.csv") - If you work in a
.Rmdfile in your manuscript folder you have to navigate up one directory level by using../: Open the file directly from the folder withread.csv("../raw-data/20200127_parlgov.csv") - Similarly, if you want to store processed data or a figure you would directly store it to the right folder.
Data Management and Data Manipulation
Getting to know the tidyverse
The R universe basically builds upon two (seemingly contradictive) approaches: base R and the tidyverse. While these two approaches are often seen as two different philosophies, they can form a symbiosis. We therefore recommend to pick whichever works best for you – or to combine the two.
Whereas base R is already implemented in R, using the tidyverse requires users to load new packages. People often find base R unintuitive and hard to read. This is why Hadley Wickham developed and introduced the tidyverse – a more intuitive approach to managing and wrangling data. Code written before 2014 was usually written in base R whereas the tidyverse style is becoming increasingly widespread. Again, which approach you prefer is rather a matter of personal taste than a decision between “right or wrong”.
The following figure shows two code chunks that are substantially identical and visualize the main differences between the two approaches.

The logic of tidyverse is fairly simple: As you can see in the graphic above, when using the tidyverse style, you start with your main object (me) and pipe (%>%) through this object by filtering, selecting, renaming, … parts of it. With these pipe operators, you read the code from left to right (or, across multiple lines, from top to bottom). In base R you would – in contrast – wrap the commands around your main object and thus read the code from the inside out.
We first familiarize ourselves with the basic logic of the tidyverse style using some examples. For this, we use some generated data.
Code: Generate data
# We use a code that is adjusted and based on examples provided here: https://declaredesign.org/r/fabricatr/articles/building_importing.html
# We set a seed to make our dataset reproducible.
set.seed(68159)
data <- fabricate(
countries = add_level(
N = 10,
month = recycle(month.abb),
gdppercapita = runif(N, min = 10000, max = 50000),
unemployment_rate = 5 + runif(N, 30,50) + ((gdppercapita > 50) * 10),
life_expectancy = 50 + runif(N, 10, 20) + ((gdppercapita > 30000) * 10)
)
)
# Drop countries
data <- data %>% select(-countries)
# We add artificial NAs
data <- data %>%
mutate(
gdppercapita = ifelse(gdppercapita > 40000 &
gdppercapita < 44000, NA, gdppercapita),
unemployment_rate = ifelse(
unemployment_rate > 50 &
unemployment_rate < 54,
NA,
unemployment_rate
)
)
# Have a look at the data
data## month gdppercapita unemployment_rate life_expectancy
## 1 Jan 31819.79 48.90126 70.19808
## 2 Feb 45485.76 58.51337 76.16142
## 3 Mar 18327.78 55.89483 67.68735
## 4 Apr 22275.86 48.61502 60.98838
## 5 May 23069.79 47.18671 66.39749
## 6 Jun 34896.12 NA 72.52299
## 7 Jul 14370.76 58.33964 66.42222
## 8 Aug NA 59.60927 74.91735
## 9 Sep NA NA 73.82523
## 10 Oct 24195.20 NA 65.26416# If your dataset becomes larger, the command "head(data)" is a useful alternative
# to get a quick glance at the data. It shows by default the first 6 rows.Filtering the data
Our dataset contains information on countries (numeric), months (characters), GDP per capita, unemployment rates, and life expectancies (all numeric). Suppose we are only interested in data from January. For this purpose, we can use the filter() function, provided by the package dplyr.1
data %>%
filter(month == "Jan")## month gdppercapita unemployment_rate life_expectancy
## 1 Jan 31819.79 48.90126 70.19808 How would you do it in base R?
data[data$month == "Jan", ]RStudio also provides shortcuts that allow you to write the pipe operator (%>%) quickly:
- Shift + Cmd + M (Mac)
- Shift + Ctrl + M (Windows)
Selecting specific variables
Sometimes, you want to select specific variables. We are interested in the unemployment rate and the months but not so much in the other variables. dplyr’s command select() allows you to do exactly this.
data %>%
select(month, unemployment_rate) %>%
head()## month unemployment_rate
## 1 Jan 48.90126
## 2 Feb 58.51337
## 3 Mar 55.89483
## 4 Apr 48.61502
## 5 May 47.18671
## 6 Jun NA How would you do it in base R?
head(data[, c("month", "unemployment_rate")])Arranging variables
Let’s say we want to know the largest unemployment rate and the lowest unemployment rate throughout the entire dataset. This can be done with the arrange() command that is added with another %>% operator to our previous code.
By default, the arrange() function sorts the data in ascending order. To display the data in descending order, we add desc().
# Arrange in ascending order
data %>%
select(month, unemployment_rate) %>%
arrange(unemployment_rate)## month unemployment_rate
## 1 May 47.18671
## 2 Apr 48.61502
## 3 Jan 48.90126
## 4 Mar 55.89483
## 5 Jul 58.33964
## 6 Feb 58.51337
## 7 Aug 59.60927
## 8 Jun NA
## 9 Sep NA
## 10 Oct NA# Arrange in descending order
data %>%
select(month, unemployment_rate) %>%
arrange(desc(unemployment_rate))## month unemployment_rate
## 1 Aug 59.60927
## 2 Feb 58.51337
## 3 Jul 58.33964
## 4 Mar 55.89483
## 5 Jan 48.90126
## 6 Apr 48.61502
## 7 May 47.18671
## 8 Jun NA
## 9 Sep NA
## 10 Oct NA How would you do it in base R?
# Arrange in ascending order
data[order(data$unemployment_rate), c("month","unemployment_rate")]
# Arrange in descending order
data[order(data$unemployment_rate, decreasing = TRUE),
c("month","unemployment_rate")]As we can see, the highest unemployment rate was in August with 59.61% and the lowest unemployment rate in May with 47.19%.
Group-wise operations
To get the highest unemployment rates by month, we use a combination of group_by() (to group our results by month) and the summarise() function.
data %>%
group_by(month) %>%
summarise(max_unemployment = max(unemployment_rate)) ## # A tibble: 10 x 2
## month max_unemployment
## <chr> <dbl>
## 1 Apr 48.6
## 2 Aug 59.6
## 3 Feb 58.5
## 4 Jan 48.9
## 5 Jul 58.3
## 6 Jun NA
## 7 Mar 55.9
## 8 May 47.2
## 9 Oct NA
## 10 Sep NA How would you do it in base R?
aggregate(data$unemployment_rate, by = list(data$month), max)In some cases, you might no longer need your variables grouped after performing transformations. In this case, you can pipe another command with %>% ungroup() at the end to ungroup your data.
Extracting unique observations
Which unique months are included in the dataset? To get this information, we use the distinct() command in dplyr. As we see, all months from January to December are present.
R allows us to also sort this data alphabetically with arrange().
# Get the distinct months in our dataset
data %>%
distinct(month)## month
## 1 Jan
## 2 Feb
## 3 Mar
## 4 Apr
## 5 May
## 6 Jun
## 7 Jul
## 8 Aug
## 9 Sep
## 10 Oct# Sort data alphabetically
data %>%
distinct(month) %>%
arrange(month)## month
## 1 Apr
## 2 Aug
## 3 Feb
## 4 Jan
## 5 Jul
## 6 Jun
## 7 Mar
## 8 May
## 9 Oct
## 10 Sep How would you do it in base R?
# Get the distinct months in our dataset
unique(data$month)
# Sort data alphabetically
sort(unique(data$month))Renaming variables
The variable name gdppercapita is hard to read. We therefore want rename it to gdp_per_capita using the rename() function from the dplyr package. The compound assignment pipe operator %<>% from the magrittr package simultaneously serves as the first pipe in a chain of commands and assigns the transformed data to the left-hand side object.
# Rename the variable "gdppercapita" to "gdp_per_capita"
data %<>%
rename(gdp_per_capita = gdppercapita) How would you do it in base R?
names(data)[names(data) == "gdppercapita"] <- "gdp_per_capita"Creating new variables
The mutate() command allows you to generate a new variable. Let’s say you want to generate a dummy that indicates if it is summer or not. We call this variable summer.
# Create a new variable called "summer"
data %>%
mutate(summer = ifelse(month %in% c("Jun", "Jul", "Aug"), 1, 0))## month gdp_per_capita unemployment_rate life_expectancy summer
## 1 Jan 31819.79 48.90126 70.19808 0
## 2 Feb 45485.76 58.51337 76.16142 0
## 3 Mar 18327.78 55.89483 67.68735 0
## 4 Apr 22275.86 48.61502 60.98838 0
## 5 May 23069.79 47.18671 66.39749 0
## 6 Jun 34896.12 NA 72.52299 1
## 7 Jul 14370.76 58.33964 66.42222 1
## 8 Aug NA 59.60927 74.91735 1
## 9 Sep NA NA 73.82523 0
## 10 Oct 24195.20 NA 65.26416 0 How would you do it in base R?
data$summer <- ifelse(data$month %in% c("Jun", "Jul", "Aug"), 1, 0)Additional features
mutate() and summarise() also have several scoped variants that allow us to apply our commands simultaneously to several variables:
_all()affects all variables_at()affects specific selected variables_if()affects conditionally selected variables
How does this work in practice? Let’s say we have good reason to believe that all NAs in our observations are 0 and we want to transform all variables with NAs at once. We then use mutate_if():
data %<>% mutate_if(is.numeric, replace_na, 0) How would you do it in base R?
data <- lapply(data, function(x) replace(x, is.na(x), 0))Data wrangling for complex data structures: A walkthrough
Following this short intro to the tidyverse, we want to showcase how sequences of data-transforming operations can be used to manage and combine complex data structures. We use a classical multi-level setup: We want to enrich multinational survey data with country-specific contextual information. For this purpose, we use data from Round 9 of the European Social Survey (ESS), a cross-national survey of nearly 20 European countries fielded between August 2018 and May 2019 and augment it with party positions from the Manifesto Project and unemployment rates from the World Bank.
Note: As shown in the examples above, a sequence of commands may be combined using a series of piped operations. For didactic purposes, we break these sequences into shorter segments in the following examples.
Election Dates
Now that we have selected and partly recoded our micro-level variables, we are confronted with a new challenge: The ESS records respondents’ recall of their voting behavior in the most recent national election. We thus need to identify the correct electoral context for each country before we can add information on party strategies. This requires that we identify the date of the most recent national election prior to a given interview.
As a first step, we need to collect information on the country-specific ESS field times. We use information on the date on which individual interviews were started, which are stored in separate variables for year, month, and day: inwyys, inwmms, and inwdds. We combine these three variables in YYYYMMDD format using the base R sprintf() command and save them as a date using the ymd() command from the tidyverse’s lubridate package. After this, we can drop the three variables from our data using dplyr’s select() command.
ess %<>%
mutate(inwdate = sprintf('%04d%02d%02d', inwyys, inwmms, inwdds)) %>%
mutate(inwdate = ymd(inwdate)) %>%
select(-inwyys,-inwmms,-inwdds)To retrieve information on country-specific field periods, we need to find the earliest and latest interview dates within each country. We therefore use the dplyr commands group_by() and summarize() to generate an auxiliary data frame named ess_dates.
ess_dates <- ess %>%
group_by(cntry) %>%
summarize(field_start = min(inwdate),
field_end = max(inwdate)) %>%
print()## # A tibble: 19 x 3
## cntry field_start field_end
## <chr> <date> <date>
## 1 AT 2018-09-18 2019-01-12
## 2 BE 2018-09-20 2019-01-27
## 3 BG 2018-11-16 2018-12-15
## 4 CH 2018-08-31 2019-02-11
## 5 CY 2018-09-18 2019-05-26
## 6 CZ 2018-11-17 2019-02-02
## 7 DE 2018-09-03 2019-03-03
## 8 EE 2018-10-01 2019-03-02
## 9 FI 2018-09-06 2019-02-18
## 10 FR 2018-10-20 2019-04-01
## 11 GB 2018-09-01 2019-02-16
## 12 HU 2019-01-31 2019-05-22
## 13 IE 2018-11-07 2019-03-31
## 14 IT 2018-12-17 2019-03-10
## 15 NL 2018-09-03 2019-01-20
## 16 NO 2018-10-04 2019-05-16
## 17 PL 2018-10-26 2019-03-20
## 18 RS 2018-09-28 2019-02-24
## 19 SI 2018-09-24 2019-02-01Now that we know the country-specific field periods, we need to find the election date preceding the respective field periods. For this purpose, we use the comprehensive collection on parliaments and elections from ParlGov. The data carries information on parties performance in both national and European parliamentary elections. As the data set comes in .csv format, we use the read.csv() command to load it into R. In order to use the data, we need to do a bit of housekeeping first:
- We use the
countrycodepackage to convert country names to ISO2C codes. This allows us to combine them with the data fromess_dateslater on. - We retain information on national parliamentary elections, thus dropping observations for elections to the European parliament.
- We select three variables we need: Country codes in ISO2C format, election dates, and unique election IDs.
- As the data set still contains many observations per election (one for each party), we use
group_by()-summarize_all()-ungroup()to collapse the data frame. As a result, it now contains one unique value forcntryandelection_datefor each value ofelection_id. - Lastly, we combine the election dates data from
pgovwith the field times fromess_dates, using the harmonizedcntryidentifiers. Usingright_join()ensure that we only keep the election dates for the 19 countries included in the right-hand side data iness_times.
pgov <- read.csv("efficient-data-r/raw-data/20200127_parlgov.csv") %>%
mutate(cntry = countrycode(country_name, "country.name", "iso2c")) %>%
filter(election_type == "parliament") %>%
select(cntry, election_date, election_id) %>%
group_by(election_id) %>%
summarize_all(unique) %>%
ungroup() %>%
right_join(ess_dates, by = "cntry")At this point, we have added all available election dates for each available country. What we want, however, is to find only the date of the most recent election preceding a country’s ESS field period. So what do we do?
- We save
election_dateas an actual date variable. This allows us to compare it to other date variables. - For each country, we exclude all elections that took place after the respective start date of the ESS field period.2
- We
group_by()countries andarrange()the observations byelection_datefrom earliest to latest. We then identify themost_recentelection that has the smallest duration between the start of the field period and the preceding election date (while we’re at it, we also store the date of previous election for each country, which will come in handy later on). We thenungroup(). - We can now use
filter()to keep only those observation whereelection_date == most_recentevaluates to true.
pgov %<>%
mutate(election_date = ymd(election_date)) %>%
filter(election_date <= field_start) %>%
group_by(cntry) %>%
arrange(election_date) %>%
mutate(most_recent = election_date[which.min(field_start - election_date)],
prev_election_date = lag(election_date)) %>%
ungroup() %>%
filter(election_date == most_recent) The rest is just a little housekeeping: We extract the years of the most recent and previous election dates and select only those variables which we are going to need later on.
Code: Election dates (housekeeping)
pgov %<>%
mutate(
election_year = year(election_date),
prev_election_year = year(prev_election_date)
) %>%
select(cntry,
election_date,
prev_election_date,
election_year,
prev_election_year)Party Positions
We now know the exact dates of the elections in which individuals cast their votes as reported in the ESS. This finally allows us to combine the survey data with information on the policy positions on which center-left parties campaigned in the respective elections. We therefore load the data set from the Manifesto Project, which comes in Stata 13+ format and therefore requires the read.dta13() command from the readStata13 package. We start with a little housekeeping, converting country names to ISO2C codes, subsetting the data to include only social democratic parties, and storing election dates as actual date variables.
Code: Manifesto Project – Initial data management
cmp <- read.dta13("efficient-data-r/raw-data/20200127_cmp2019b.dta") %>%
mutate(cntry = countrycode(countryname, "country.name", "iso2c")) %>%
filter(parfam == "soc social democratic") %>%
mutate(election_date = as.Date(edate))
We then right_join() the data frame pgov to the Manifesto data in cmp, matching observations by both cntry and election_date, and select all relevant variables. Next to pervote, which contain parties’ vote shares in the most recent election, we keep rile and welfare. The former is parties’ general left-right position, the latter their position on welfare issues.
cmp %<>%
right_join(pgov, by = c("cntry", "election_date")) %>%
select(
cntry,
election_date,
election_year,
prev_election_date,
prev_election_year,
party,
partyname,
partyabbrev,
pervote,
rile,
welfare
)One problem we still need to address is that some countries have multiple parties of the social democratic party family. For the sake of simplicity, we choose to focus only on one party per country, namely the strongest. Therefore, we again group by country and keep only those observations with the highest vote percentage. We now have a data set that contains the positions of each countries’ main center-left party, based on the most recent election preceding the ESS, which we can easily merge with our ESS survey data using the cntry identifier.
cmp %<>%
group_by(cntry) %>%
filter(pervote == max(pervote, na.rm = TRUE)) %>%
ungroup()
## View data
cmp %>%
select(cntry, election_year, partyname, partyabbrev, welfare)
## Add to ESS
ess %<>% left_join(cmp, by = "cntry")Unemployment Rates
Now, suppose we also want to retrieve information on the average national unemployment rate during the legislative period preceding the most recent national election. For this purpose, we use data on annual national unemployment rates from the World Bank.
Code: World Bank unemployment rates – Initial data management
unemp <-
read.csv("efficient-data-r/raw-data/20200114_worldbank_unemprate.csv",
skip = 4L) %>%
mutate(cntry = countrycode(Country.Name, "country.name", "iso2c")) %>%
filter(cntry %in% cmp$cntry)
Following some initial data management, we can see that the data is in wide format: Every country has only one row while their annual unemployment rates are stored in multiple columns named X1960, …, X2019. So what do we do?
First, we select our country-identifier along with all yearly observations starting with X (except an unspecified additional variable named X). We then use the melt() command from the reshape2 package. This changes the data format from wide to long: We now have 60 observations per country (for years 1960-2019), uniquely identified by a new variable with we rename to year, with their corresponding values stored in the new variable we rename to unemprate. The year variable contains the previous variable names (X1960, …, X2019). After omitting the leading X character, for which we use the base R substr() command, we store the years as numeric values.
unemp %<>%
select(cntry, starts_with("X"),-X) %>%
melt(id.vars = "cntry") %>%
rename(year = variable,
unemprate = value) %>%
mutate(year = as.numeric(substr(year, 2L, 5L))) Using the long-format data, we then proceed as follows:
- We select the country-specific information in the most recent and previous election years from the
pgovdata frame, andleft_join()it with theunempdata frame using our country identifiers. Thereby, we supplement each of the 60 yearly observations per country with the information on the relevant time span. - After grouping the data set by
cntry, we can use this information tofilter()only the years of each country’s past legislative period. - The
summarize()function, lastly, allows us to average across these observation to retrieve a single observation per country. This gives us the desired average unemployment rate during the past legislative period, which we may merge with the survey data using thecntryidentifier.
unemp %<>%
left_join(pgov %>% select(cntry, election_year, prev_election_year),
by = c("cntry")) %>%
group_by(cntry) %>%
filter(year >= prev_election_year &
year <= election_year) %>%
summarize(unemprate = mean(unemprate))
ess %<>% left_join(unemp, by = "cntry")Programming and Debugging
Identifying center-left parties in the ESS
Now we need a variable which indicates whether a respondent of the ESS voted for a center-left party. Unfortunately, the variables indicating the party a respondent voted for at the last election are rather messy: Each country has its own variable and party names are not consistent: sometimes, the ESS uses abbreviations, sometimes full names. Thus, we need to check the ESS codebook and the ESS Round 9 Appendix on Political Parties to manually code the names of center-left parties.
To save the information, we generate a new auxiliary data frame which we use later on.
Code: Preparation of auxiliary data
## Main center-left parties
aux <- list(
AT = "SPÖ",
BE = "SP.A",
BG = "Balgarska sotsialisticheska partiya (BSP)" ,
CH = "Social Democratic Party / Socialist Party" ,
CY = "Progressive Party of Working People (AKEL)" ,
CZ = "ČSSD",
DE = "SPD" ,
EE = "Sotsiaaldemokraatlik Erakond",
FI = "Social Democratic Party",
FR = "PS (Parti Socialiste)",
GB = "Labour",
HU = "MSZP (Magyar Szocialista Párt)",
IE = "Labour",
IT = "Partido Democratico (PD)",
NL = "Socialist Party",
NO = "Arbeiderpartiet",
PL = "Zjednoczona Lewica",
RS = "Boris Tadic, Cedomir Jovanovic - Savez za bolju Srbiju - LDP, LSV, SDS",
SI = "L - Levica"
) %>%
melt() %>%
rename(
ess_party_name = value,
cntry = L1
)
## Match vote variables and country codes
vote_var <- ess %>%
select(starts_with("prtv"),-prtvede1) %>%
names()
vote_var_order <-
sapply(aux$cntry, function (x)
which(grepl(x, toupper(vote_var))))
## Add to auxiliary data frame
aux %<>%
mutate(vote_var = vote_var[vote_var_order]) %>%
mutate_if(is.factor, as.character)Programming a function
Now, we want to add a variable that indicates whether a respondent voted for a center left party (as opposed to voting for a different party or not voting at all). To this end, we program a function named ess_center_left(). This function takes three input arguments. micro_data, aux_data, and group_var. micro_data is a data frame containing the survey data, i.e., our ess object. aux_datais a data frame with auxiliary information, i.e., our aux object. group_var, lastly, is a character string containing the name of the grouping variable (cntry) in both data frames. After several operations which we discuss in detail below, the function returns the modified data frame micro_data, now with an added variable v_center_left.
ess_center_left <- function(micro_data, aux_data, group_var) {
## A priori consistency checks
...
## Main function
...
## A posteriori consistency checks
...
## Return output
return(micro_data)
}So what goes into our function body? We divide our function into three parts:
- A priori consistency checks
- The main function
- A posteriori consistency check
In the following, we will walk through each part separately and subsequently combine them into one function.
A priori consistency checks
A priori consistency checks test whether we supply the correct inputs to our function. In a first step, we check if both micro_data and aux_data actually contain a variable of the name specified in group_var. If either of them does not, the function stops and returns an error message. In a second step, also check for the unique values (i.e., country codes) of group_var in micro_data and aux_data and establish their intersection in group_vals_both. The third step establishes which, if any, values of group_var are missing in the micro and auxiliary data. In the fourth and final step, we print corresponding warning messages that inform us which countries are not included in both the micro and auxiliary data. For these, the new variable v_center_left will not be created.
# 1) Check if group_var is a variable in micro_data and aux_data
if (not(group_var %in% names(micro_data))) {
stop("group_var is not a variable in micro_data.")
} else if (not(group_var %in% names(aux_data))) {
stop("group_var is not a variable in aux_data.")
} else {
# 2) Unique values of group_var
group_vals_micro <- unique(micro_data[, group_var])
group_vals_aux <- unique(aux_data[, group_var])
group_vals_both <- group_vals_micro[group_vals_micro %in% group_vals_aux]
# 3) Missing values of group_var in micro_data and aux_data
group_vals_only_micro <-
group_vals_micro[not(group_vals_micro %in% group_vals_both)]
group_vals_only_aux <-
group_vals_aux[not(group_vals_aux %in% group_vals_both)]
# 4) Missing group_var values in micro_data or aux_data
if (length(group_vals_only_micro) > 0) {
warning(paste("Group values only in micro_data:",
group_vals_only_micro, sep = " "))
}
if (length(group_vals_only_aux) > 0) {
warning(paste("Group values only in aux_data:",
group_vals_only_aux, sep = " "))
}
}Main function
If our data inputs pass the initial consistency checks, the function generates the new variable v_center_left within each available country. We begin by defining an auxiliary identifier, aux_id that allows us to uniquely identify observations in the ESS data. This will allow us to merge the newly created variable to the original data later on. We then generate a list, vote_recoded that serves a container for the country-specific data that we generate in the subsequent loop through the different countries.
In this loop, we first retrieve the name of the ESS vote choice variable and the name of the center-left party in a given country. We then subset the data to observations from this country and retain three variables only: aux_id, vote (i.e., whether someone voted or not), and the country-specific vote choice variable, which we subsequently rename to vote_choice and store in our system’s native encoding.
Following this, we assign one out of four possible values to the new variable v_center_left:
- If we have no information on respondents’ voting behavior:
NA - If the respondent did not vote or was not eligible to vote:
Did not vote - If the respondent voted for the country’s main center-left party:
Yes - If the respondent voted for a different party:
No
After this, we collapse the list of country-specific data frames to a single data frame which we then add to the original data frame micro_data by our identifier aux_id.
# Auxiliary ID
micro_data$aux_id <- seq_len(nrow(micro_data))
# List container for data frames containing v_center_left
vote_recoded <- list()
# Loop through groups
for (j in group_vals_both) {
# Name of the group's vote choice variable
vote_var_j <- aux_data$vote_var[aux_data$cntry == j]
# Name of the group's center left party (in native encoding)
ess_party_name_j <- aux_data$ess_party_name[aux_data$cntry == j]
# Generate v_center_left for this group
vote_recoded[[j]] <- micro_data %>%
filter(cntry == j) %>% # subset to group
select(aux_id, vote, vote_var_j) %>% # select variables
rename(vote_choice = !!as.name(vote_var_j)) %>% # rename vote choice
mutate(vote_choice = enc2native(vote_choice), # harmonize encoding
v_center_left = # create v_center_left
ifelse(
is.na(vote) & is.na(vote_choice),
NA, # missing information
ifelse(
vote %in% c("No", "Not eligible to vote"),
"Did not vote", # non-voters
ifelse(vote_choice == ess_party_name_j,
"Yes", # center-left voters
"No") # voted for other party
)
))
}
# Collapse list of data frames to a single data frame
vote_recoded %<>%
bind_rows() %>%
select(aux_id, v_center_left)
# Remove old versions of v_center_left if present
if ("v_center_left" %in% names(micro_data)) {
micro_data %<>% select(-v_center_left)
}
# Add new variable to micro_data by aux_id
micro_data %<>%
full_join(vote_recoded, by = "aux_id") %>% # add new variable
select(-aux_id) # drop auxiliary IDA posteriori consistency checks
After generating our new variable v_center_left within each country, we want to know if our function actually worked. For this purpose, we check whether any country has zero percent of center-left voters. Given that we supplied a name for one center-left party per country, this would likely suggest a mismatch in our inputs.
# Proportions of center left voters within each group
sample_prop <- micro_data %>%
select(group_var, v_center_left) %>%
group_by(!!as.name(group_var)) %>%
summarize(prop_v_center_left = mean(v_center_left == "Yes", na.rm = TRUE))
# Check if any group has 0% center left voters
if (any(sample_prop$prop_v_center_left == 0)) {
warning(paste(
"No center-left voters in",
paste(sample_prop$cntry[sample_prop$prop_v_center_left == 0],
collapse = " "),
"- check your inputs!",
sep = " "
))
}Now, we can put the parts together and define our function.
Code: Full function
ess_center_left <- function(micro_data, aux_data, group_var) {
## A priori consistency checks
# 1) Check if group_var is a variable in micro_data and aux_data
if (not(group_var %in% names(micro_data))) {
stop("group_var is not a variable in micro_data.")
} else if (not(group_var %in% names(aux_data))) {
stop("group_var is not a variable in aux_data.")
} else {
# 2) Unique values of group_var
group_vals_micro <- unique(micro_data[, group_var])
group_vals_aux <- unique(aux_data[, group_var])
group_vals_both <- group_vals_micro[group_vals_micro %in% group_vals_aux]
# 3) Missing values of group_var in micro_data and aux_data
group_vals_only_micro <-
group_vals_micro[not(group_vals_micro %in% group_vals_both)]
group_vals_only_aux <-
group_vals_aux[not(group_vals_aux %in% group_vals_both)]
# 4) Missing group_var values in micro_data or aux_data
if (length(group_vals_only_micro) > 0) {
warning(paste("Group values only in micro_data:",
group_vals_only_micro, sep = " "))
}
if (length(group_vals_only_aux) > 0) {
warning(paste("Group values only in aux_data:",
group_vals_only_aux, sep = " "))
}
}
## Main function
# Auxiliary ID
micro_data$aux_id <- seq_len(nrow(micro_data))
# List container for data frames containing v_center_left
vote_recoded <- list()
# Loop through groups
for (j in group_vals_both) {
# Name of the group's vote choice variable
vote_var_j <- aux_data$vote_var[aux_data$cntry == j]
# Name of the group's center left party (in native encoding)
ess_party_name_j <- aux_data$ess_party_name[aux_data$cntry == j]
# Generate v_center_left for this group
vote_recoded[[j]] <- micro_data %>%
filter(cntry == j) %>% # subset to group
select(aux_id, vote, vote_var_j) %>% # select variables
rename(vote_choice = !!as.name(vote_var_j)) %>% # rename vote choice
mutate(vote_choice = enc2native(vote_choice), # harmonize encoding
v_center_left = # create v_center_left
ifelse(
is.na(vote) & is.na(vote_choice),
NA, # missing information
ifelse(
vote %in% c("No", "Not eligible to vote"),
"Did not vote", # non-voters
ifelse(vote_choice == ess_party_name_j,
"Yes", # center-left voters
"No") # voted for other party
)
))
}
# Collapse list of data frames to a single data frame
vote_recoded %<>%
bind_rows() %>%
select(aux_id, v_center_left)
# Remove old versions of v_center_left if present
if ("v_center_left" %in% names(micro_data)) {
micro_data %<>% select(-v_center_left)
}
# Add new variable to micro_data by aux_id
micro_data %<>%
full_join(vote_recoded, by = "aux_id") %>% # add new variable
select(-aux_id) # drop auxiliary ID
## A posteriori consistency checks
# Proportions of center left voters within each group
sample_prop <- micro_data %>%
select(group_var, v_center_left) %>%
group_by(!!as.name(group_var)) %>%
summarize(prop_v_center_left = mean(v_center_left == "Yes", na.rm = TRUE))
# Check if any group has 0% center left voters
if (any(sample_prop$prop_v_center_left == 0)) {
warning(paste(
"No center-left voters in",
paste(sample_prop$cntry[sample_prop$prop_v_center_left == 0],
collapse = " "),
"- check your inputs!",
sep = " "
))
}
## Return output
return(micro_data)
}Debugging
Now we apply this function:
ess %<>%
ess_center_left(aux_data = aux, group_var = "cntry")## Note: Using an external vector in selections is ambiguous.
## i Use `all_of(vote_var_j)` instead of `vote_var_j` to silence this message.
## i See <https://tidyselect.r-lib.org/reference/faq-external-vector.html>.
## This message is displayed once per session.## Note: Using an external vector in selections is ambiguous.
## i Use `all_of(group_var)` instead of `group_var` to silence this message.
## i See <https://tidyselect.r-lib.org/reference/faq-external-vector.html>.
## This message is displayed once per session.## Warning in ess_center_left(., aux_data = aux, group_var = "cntry"): No
## center-left voters in DE - check your inputs!There seems to be something wrong with Germany. Let’s have a look:
ess %>%
filter(cntry == "DE") %>%
select(prtvede2) %>%
table()## .
## Alliance 90/The Greens (Bündnis 90/Die Grünen)
## 289
## Alternative for Germany (AFD)
## 111
## Christian Democratic Union/Christian Social Union (CDU/CSU)
## 558
## Free Democratic Party (FDP)
## 147
## National Democratic Party (NPD)
## 2
## Other
## 36
## Pirate Party (Piratenpartei)
## 4
## Social Democratic Party (SPD)
## 355
## The Left (Die Linke)
## 125aux %>%
filter(cntry == "DE")## ess_party_name cntry vote_var
## 1 SPD DE prtvede2As we can see, we do have a sizable number of center-left voters in Germany. However, the correct code for the SPD is not “SPD” but “Social Democratic Party (SPD)”. So let’s fix this bug and apply the function again:
aux$ess_party_name[aux$cntry == "DE"] <- "Social Democratic Party (SPD)"
# Apply the function
ess %<>%
ess_center_left(aux_data = aux, group_var = "cntry")Lastly, we save the data in the gen-data folder:
save(ess, file = "efficient-data-r/gen-data/ess-proc.RData")Working with the data
We can now use the data to approach our initial question through some simple descriptives. We first generate a binary version of our outcome variable v_center_left, where Yes equals TRUE whereas both No and Did not vote equal FALSE. We then split the data into a list of 19 country-specific data frames. Using sapply(), we then apply the a function for estimation and prediction to each of 19 data sets. Specifically, we run binary logistic regressions of v_center_left on uemp5yr. We then use the predict() function to retrieve the predicted probabilities of center-left votes among the unemployed along with their 95% confidence intervals for each country.
ess_est <- ess %>%
mutate(v_center_left = (v_center_left == "Yes")) %>%
group_split(cntry) %>%
lapply(function(dat) {
mod <- glm(v_center_left ~ uemp5yr,
data = dat,
weights = dweight)
pred <- predict(mod,
newdata = data.frame(uemp5yr = "Yes"),
se.fit = TRUE)
pos_welfare <- unique(dat$welfare)
output <- data.frame(
est = pred$fit,
lower95 = pred$fit + qnorm(.025) * pred$se,
upper95 = pred$fit + qnorm(.975) * pred$se,
welfare = unique(dat$welfare)
)
return(output)
}) %>%
bind_rows()We can then plot the relationship between center-left support among the unemployed and center-left parties’ welfare policy positions. As we can see below, based on 14 out of the 19 countries for which data on party positions is available, there does not seem to be a straightforward relationship between the two variables.
ess_est %>%
ggplot(aes(x = welfare, y = est)) +
geom_errorbar(aes(ymin = lower95, ymax = upper95)) +
geom_point() +
labs(x = "Center Left Welfare Policy Position",
y = "Proportion of Unemployed Voting for Center Left")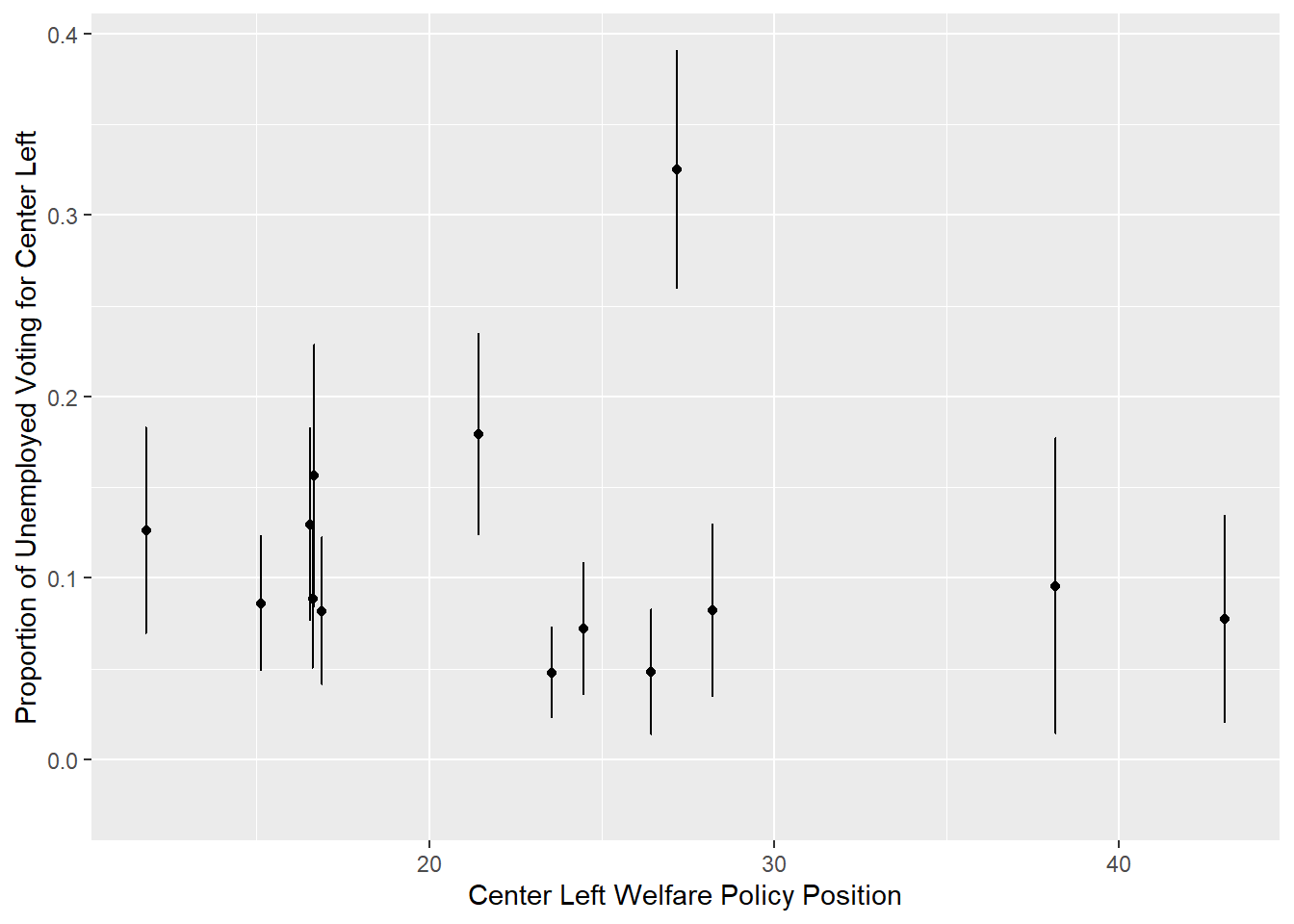
About the Presenters
Denis Cohen is a postdoctoral fellow in the Data and Methods Unit at the Mannheim Centre for European Social Research (MZES), University of Mannheim, and one of the organizers of the MZES Social Science Data Lab. His research focus lies at the intersection of political preference formation, electoral behavior, and political competition. His methodological interests include quantitative approaches to the analysis of clustered data, measurement models, data visualization, strategies for causal identification, and Bayesian statistics.
Cosima Meyer is a doctoral researcher and lecturer at the University of Mannheim and one of the organizers of the MZES Social Science Data Lab. Motivated by the continuing recurrence of conflicts in the world, her research interest on conflict studies became increasingly focused on post-civil war stability. In her dissertation, she analyzes leadership survival – in particular in post-conflict settings. Using a wide range of quantitative methods, she further explores questions on conflict elections, women’s representation as well as autocratic cooperation.
Marcel Neunhoeffer is a PhD Candidate and Research Associate at the chair of Political Science, Quantitative Methods in the Social Sciences, at the University of Mannheim. His research focuses on political methodology, specifically on the application of deep learning algorithms to social science problems. His substantive interests include data privacy, political campaigns, and forecasting elections.
Oliver Rittmann is a PhD Candidate and Research Associate at the chair of Political Science, Quantitative Methods in the Social Sciences, at the University of Mannheim. His research focuses on legislative studies and political representation. His methodological expertise includes statistical modeling, authomated text and video analysis, and subnational public opinion estimation.
Further Reading
- Bryan, Jenny and Jim Hester. Happy Git and GitHub for the useR.
- Grolemund, Garrett. A Tidyverse Cookbook.
- Grolemund, Garrett and Hadley Wickham. R for Data Science.
- Healy, Kieran. Data Visualization: A Practical Introduction.
- Wickham, Hadley. Advanced R.
- Base R Cheat Sheet.
- Data Transformation with dlyr Cheat Sheet.
{By default, when several packages use identical names for different functions, R and R Studio use the package that was loaded last. This masks functions of the same name from packages loaded before. The sequence of loading your packages in R is therefore important. It is good practice to add a prefix with the required package before each function; i.e.
packagename::function(). Alternatively, the packageconflictedis helpful – it makes every package conflict an error and tells you which package with the same functions are available. You can then identify the package that you want or need to use. This way, you make sure that you do not accidentally use a function from a different package without recognizing it.}↩︎{Additional problems could arise if a national election had taken place during the ESS field period. To identify such cases, we could simple create a true/false indicator variable using
mutate(flag = election_date >= field_start & election_date <= field_end).}↩︎
