Applied Bayesian Statistics Using Stan and R
Whether researchers occasionally turn to Bayesian statistical methods out of convenience or whether they firmly subscribe to the Bayesian paradigm for philosophical reasons: The use of Bayesian statistics in the social sciences is becoming increasingly widespread. However, seemingly high entry costs still keep many applied researchers from embracing Bayesian methods. Next to a lack of familiarity with the underlying conceptual foundations, the need to implement statistical models using specific programming languages remains one of the biggest hurdles. In this Methods Bites Tutorial, Denis Cohen provides an applied introduction to Stan, a platform for statistical modeling and Bayesian statistical inference.
Readers will learn about:
- fundamental concepts in Bayesian statistics
- the Stan programming language
- the R interface RStan
- the workflow for Bayesian model building, inference, and convergence diagnosis
- additional R packages that facilitate statistical modeling using Stan
Through numerous applied examples, readers will also learn how to write and run their own models.
This blog post is based on Denis’ workshop in the MZES Social Science Data Lab in Spring 2019. The original workshop materials can be found on our GitHub.
Contents
Setup
Setting up Stan and its R interface RStan can be somewhat time-consuming as it requires the installation of a C++ compiler. Readers should follow these instructions on the Stan Development Team’s GitHib to install and configure the rstan package and its prerequisites on their operating system.
The installation of some additional packages is necessary for working through this tutorial and for exploring further opportunities for applied Bayesian modeling using RStan.
Code: Packages used in this tutorial
## Packages
pkgs <- c("rstan", "bayesplot", "shinystan", "coda", "dplyr")
## Install uninstalled packages
lapply(pkgs[!(pkgs %in% installed.packages())], install.packages)
## Load all packages to library
lapply(pkgs, library, character.only = TRUE)Stan
What Is Stan?
In the words of the developers,
Stan is a state-of-the-art platform for statistical modeling and high-performance statistical computation. Thousands of users rely on Stan for statistical modeling, data analysis, and prediction in the social, biological, and physical sciences, engineering, and business. Users specify log density functions in Stan’s probabilistic programming language and get:
- full Bayesian statistical inference with MCMC sampling (NUTS, HMC)
- approximate Bayesian inference with variational inference (ADVI)
- penalized maximum likelihood estimation with optimization (L-BFGS)
Source: https://mc-stan.org/
Why Stan?
Stan is an open-source software that provides an intuitive language for statistical modeling along with fast and stable algorithms for fully Bayesian inference. The software offers high flexibility with only few limitations. The development process is highly transparent and publicly documented on the Stan Development Repository on GitHub.
Users benefit from various helpful resources. This includes, firstly, the extensive official documentation of Stan’s functionality in the User’s Guide, Language Reference Manual, and Language Functions Reference. Secondly, users benefit from a large and active online community in the Stan Forums and on Stack Overflow, where many members of the Stan Development Team regularly address users’ questions and troubleshoot their code. Additionally, an expanding collection of case studies, tutorials, papers and textbooks offer valuable inputs for both new and experienced users.
The Stan language is compatible with various editors for syntax highlighting, formatting, and checking (incl. RStudio and Emacs).
RStan and Other Stan Interfaces
Stan is interfaced with numerous softwares, including
- RStan (R)
- PyStan (Python)
- CmdStan (shell, command-line terminal)
- MatlabStan (MATLAB)
- Stan.jl (Julia)
- StataStan (Stata)
- MathematicaStan (Mathematica)
- ScalaStan (Scala)
While this blog post illustrates the use of the R interface RStan, users with other preferences may use the corresponding interface to call Stan from their preferred software.
Aside from the widespread popularity of R for programming, statistical computing and graphics, one of the primary reasons why we focus on RStan in this blog post is the availability of a broad range of packages that facilitate the use of RStan for applied researchers. These include
- rstan: General R Interface to Stan
- shinystan: Interactive Visual and Numerical Diagnostics and Posterior Analysis for Bayesian Models
- bayesplot: Plotting functions for posterior analysis, model checking, and MCMC diagnostics.
- brms: Bayesian Regression Models using ‘Stan’, covering a growing number of model types
- rstanarm: Bayesian Applied Regression Modeling via Stan, with an emphasis on hierarchical/multilevel models
- edstan: Stan Models for Item Response Theory
- rstantools: Tools for Developing R Packages Interfacing with ‘Stan’
Bayesian Fundamentals
We start our discussions of the fundamental concepts of Bayesian statistics and inference with the following excerpt:
In the Bayesian world the unobserved quantities are assigned distributional properties and, therefore, become random variables in the analysis. These distributions come in two basic flavors. If the distribution of the unknown quantity is not conditioned on fixed data, it is called prior distribution because it describes knowledge prior to seeing data. Alternatively, if the distribution is conditioned on data that we observe, it is clearly updated from the unconditioned state and, therefore, more informed. This distribution is called posterior distribution. […] The punchline is this: All likelihood-based models are Bayesian models in which the prior distribution is an appropriately selected uniform prior, and as the size of the data gets large they are identical given any finite appropriate prior. So such empirical researchers are really Bayesian; they just do not know it yet.
Source: Gill and Witko (2013)
As we have just learned, Bayesians, contrary to the frequentist paradigm, treat data as known and fixed, whereas parameters are considered unknown random quantities. Bayesians express their knowledge about parameters distributionally. To build some intuition about this, we consider the classical example of a series of coin tosses below. In doing so, we introduce and discuss three fundamental concepts in Bayesian statistics: The likelihood, the prior distribution, and the posterior distribution. Readers already familiar with these concepts may continue reading here.
Likelihood function
A likelihood function is an assignment of a parametric form for the data, \(\mathbf{y}\). This involves stipulating a data generating process, where we specify a probability density function (pdf) or probability mass function (pmf) governed by a (set of) parameter(s) \(\theta\). This is commonly denoted \(p(\mathbf{y}|\theta)\). Given \(\theta\), one can specify the relative likelihood of observing a given value \(y\). Treating the observed values \(\mathbf{y}\) as given yields the logical inversion “which unknown \(\theta\) most likely produces the known data \(\mathbf{y}\)?”. This is what we call the likelihood function, often denoted \(L(\theta | \mathbf{y})\). In practice, Bayesian practitioners often use the notations \(p(\mathbf{y}|\theta)\) and \(L(\theta | \mathbf{y})\) interchangeably. Here, we will stick to the former.
How can we think of the likelihood function in our present example? First, we need to think about the data generating process that produces the results of our series of coin flips. Coin flips produce a series of binary outcomes, i.e., a series of heads and tails. We can think of these as realizations of a series of Bernoulli trials following a binomial distribution. The binomial distribution characterizes a series of independent realizations of Bernoulli trials, where a probability parameter \(\pi\) governs the number of successes, \(k\), out of the total number of trials, \(n\). It is this probability parameter \(\pi\) that characterizes the fairness of the coin. For example, a value of \(\pi=0.5\) would indicate a perfectly fair coin, equally likely to produce heads or tails.
As \(\pi\) determines the probability mass of \(k\) for any number of flips, \(n\), in the coin flip example, we can think of it as the equivalent to what we referred to as \(\theta\) in the general notation above. When we flip a coin \(n\) times and observe \(k\) heads (and, thus, \(n-k\) tails), we can think of the data as the number of heads, \(k\), given the total number of flips, \(n\). This is the equivalent to what we referred to as \(\mathbf{y}\) in the general notation. So given a fixed number of flips, \(k\) is generated according to a binomial distribution governed by the parameter \(\pi\): \(k \sim \text{Binomial}(n, \pi)\). The parameter \(\pi\), then, is our unknown quantity of interest.
Prior distribution
The prior distribution is a distributional characterization of our belief about a parameter prior to seeing the data. We denote the corresponding probability function as \(p(\theta)\). The prior distribution can be either (1) substantively informed by previous research or expert assessment, (2) purposefully vague, and thus, rather uninformative, or (3) weakly informative, purposefully assigning low density for unreasonable value ranges of a given parameter without otherwise conveying substantive information. Specifying a prior distribution includes statements about the distribution’s family, density, and support.
What does this mean in the context of our example? Prior to first flipping the coin, we may have a (more or less specific) belief about the parameter \(\pi\) that characterizes the fairness of the coin. In absence of evidence that suggests otherwise, we would likely think that the coin was fair, though we would probably also reserve some skepticism and acknowledge that the coin might in fact not be fair. So how can this belief be expressed distributionally?
We know that \(\pi\) is a probability parameter. Probabilities cannot lie outside of the unit interval, i.e., \(\pi \in [0, 1]\). This defines the support of our prior distribution. We also know that probabilities can take any real number inside the unit interval. So we need a probability density function that governs the relative likelihood of \(\pi\) for every possible value inside the unit interval. The beta distribution is an ideal candidate because it ticks both boxes, offering a continuous probability distribution over the unit interval. This gives us the family of our prior distribution.
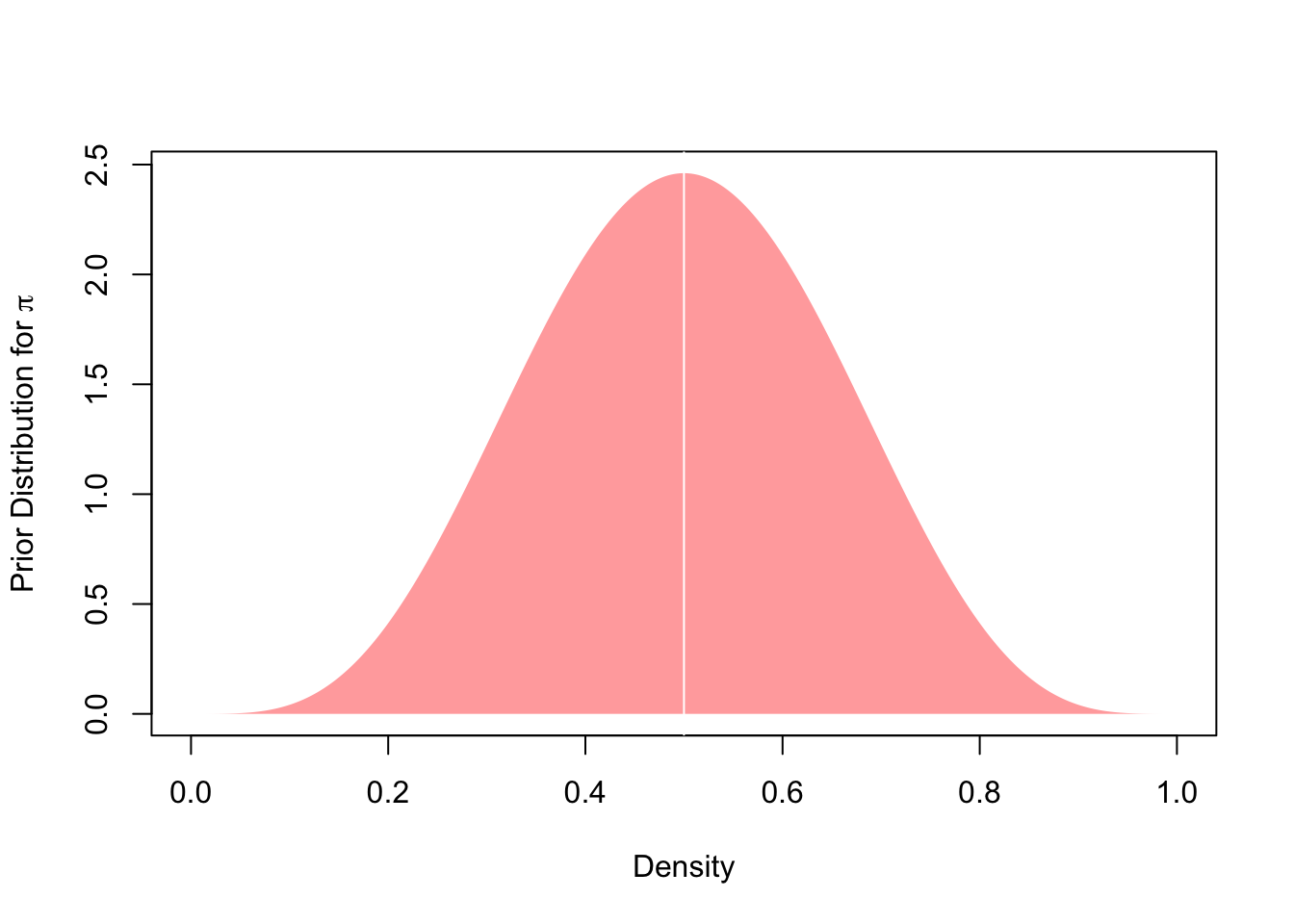
This leaves us with the density. After all, we have a weakly informative belief that the coin is more likely to be fair than it is to be (extremely) unfair. In other words, we want our prior probability density of \(\pi\) to be highest at the value \(0.5\) and lower the farther we move away from \(0.5\). Given our support, the probability will be deterministically equal to zero for values \(\pi<0\) and \(\pi>1\). So how do we get there? The trick is to specify suitable hyperparameters for the beta distribution that governs the prior probability density of our parameter \(\pi\). As the beta distribution is defined by two shape parameters, \(\alpha\) and \(\beta\), we need to choose values for the two that yield a distributional shape that conforms to our prior belief. As we can see below, \(\alpha = 5\) and \(\beta = 5\) are suitable candidates:
Code: Defining and plotting the prior distribution
len.pi <- 1001L ### number of candidate values for pi
pi <- seq(0, 1, length.out = len.pi) ### candidate values for pi
a <- b <- 5 ### hyperparameters
prior <- dbeta(pi, a, b) ### prior distribution
## Plot
plot( ### set up empty plot, specify labels
pi, prior,
type = 'n',
xlab = "Density",
ylab = expression(paste("Prior Distribution for ", pi))
)
polygon( ### draw density distribution
c(rep(0, length(pi)), pi),
c(prior, rev(prior)),
col = adjustcolor('red', alpha.f = .4),
border = NA
)
abline( ### add vertical at pi = 0.5
v = .5,
col = 'white'
)
Posterior distribution
The posterior distribution results from updating our prior belief about the parameter(s), \(p(\theta)\), through the observed data included in the likelihood function, \(p(\mathbf{y}|\theta)\). It thus yields our distributional belief about \(\theta\) given the data: \(p(\theta | \mathbf{y})\). The underlying calculation follows the proportional version of Bayes’ Law: \(p(\theta | \mathbf{y}) \propto p(\theta) \times p(\mathbf{y}|\theta)\). By multiplying the prior probability density function and the likelihood function, we get the posterior probability function of \(\theta\). It provides us with a weighthed combination of likelihood and prior: The prior pulls the posterior density toward the center of gravity of the prior distribution, but as the data grows large, the likelihood becomes increasingly influential and eventually dominates the prior.
What does this mean in the context of our example of a series of coin flips? As discussed above, we start out with our prior belief summarized by the following beta distribution: \(p(\pi) \sim \text{beta}(\alpha=5,\beta=5)\). After every coin flip \(i=1,...,n\), we update our belief about \(\pi\) by multiplying the prior pdf with the probability mass function of the binomial distribution, which is given by \(p(n,k|\pi) = {n \choose k} \pi^k (1-\pi)^{(n-k)}\). As \(n\) grows large, this latter component becomes increasingly influential in determining the posterior distribution.
As the beta distribution is conjugate to the binomial likelihood (i.e., in the same probability distribution family), the resulting posterior distribution, \(p(\pi|n,k)\), will also be a beta distribution with updated hyperparameters \(\alpha^{\prime}\) and \(\beta^{\prime}\): \(p(\pi|n,k) \sim \text{beta}(\alpha^{\prime}=\alpha+k,\beta^{\prime}=\beta+n-k)\). These updated hyperparameters then determine the probability density of the resulting posterior distribution after every additional coin flip of the series.
Example: Flipping a Coin 200 Times
Suppose we flip a coin up to 200 times. Unbeknownst to us, the coin is far from fair – it is on average four times as likely to produce heads as it is to produce tails – that is, \(\pi=0.8\). We slowly learn about this in the process of flipping the coin repeatedly, keeping score of the number of flips \(n\) and the number of heads \(k\) after each flip. This is called Bayesian updating.
The code below implements this experiment. It simulates a series of \(n=200\) coin flips and records the number of heads \(k_i\) at every \(i\)th flip. Based on this information, it retrieves the analytical solutions for the posterior mean along with its 95% credible interval at every turn.
Code: Simulating the experiment
set.seed(20190417) ### set seed for replicability
len.pi <- 1001L ### number of candidate values for pi
pi <- seq(0, 1, length.out = len.pi) ### candidate values for pi
a <- b <- 5 ### hyperparameters
n <- 200 ### num. of coin flips
pi_true <- .8 ### true parameter
data <- rbinom(n, 1, pi_true) ### n coin flips
posterior <- matrix(NA, 3L, n) ### matrix container for posterior
for (i in seq_len(n)) {
current.sequence <- data[1:i] ### sequence up until ith draw
k <- sum(current.sequence) ### number of heads in current sequence
##### Updating
a.prime <- a + k
b.prime <- b + i - k
### Analytical means and credible intervals
posterior[1, i] <- a.prime / (a.prime + b.prime)
posterior[2, i] <- qbeta(0.025, a.prime, b.prime)
posterior[3, i] <- qbeta(0.975, a.prime, b.prime)
}
## Plot
plot( ### set up empty plot with labels
1:n, 1:n,
type = 'n',
xlab = "Number of Coin Flips",
ylab = expression(
paste(
"Posterior Means of ",
pi, " (with 95% Credible Intervals)",
sep = " "
)
),
ylim = c(0, 1),
xlim = c(1, n)
)
abline( ### reference line for the true pi
h = c(.5, .8),
col = "gray80"
)
rect(-.5, qbeta(0.025, 5, 5), ### prior mean + interval at i = 0
0.5, qbeta(0.975, 5, 5),
col = adjustcolor('red', .4),
border = adjustcolor('red', .2))
segments(-.5, .5,
0.5, .5,
col = adjustcolor('red', .9),
lwd = 1.5)
polygon( ### posterior means + intervals
c(seq_len(n), rev(seq_len(n))),
c(posterior[2, ], rev(posterior[3, ])),
col = adjustcolor('blue', .4),
border = adjustcolor('blue', .2)
)
lines(
seq_len(n),
posterior[1, ],
col = adjustcolor('blue', .9),
lwd = 1.5
)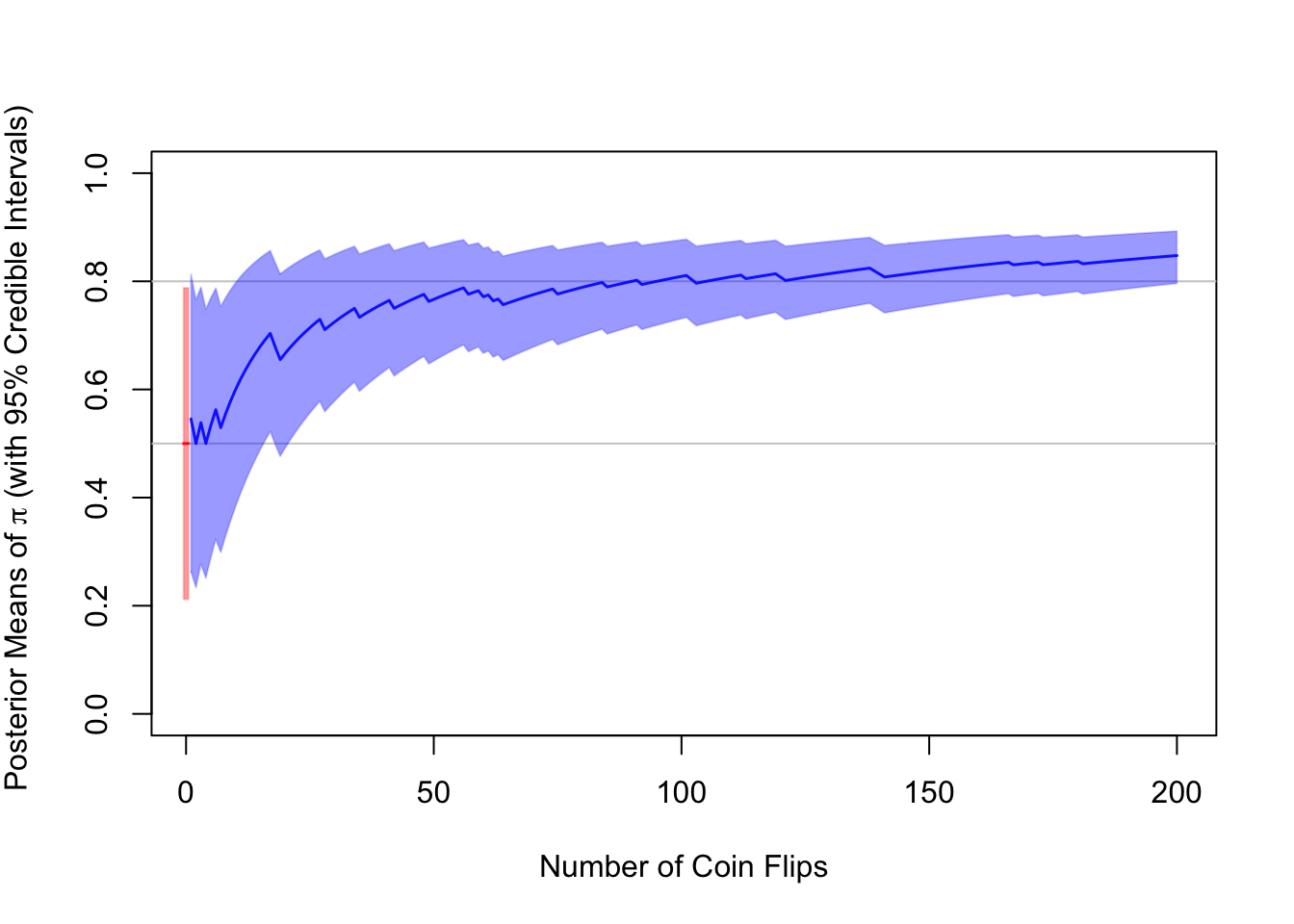
The plot above shows the prior distribution with its 95% credible interval at \(i=0\) (in red) and the updated posterior distributions with their 95% credible intervals at every coin flip \(i=1,...,n\). As we can see, even after just a couple of coin flips, the posterior distribution departs from the center of gravity of the prior distribution and converges toward the proportion of heads, \(\frac{k}{n}\), in the data. After \(n=200\) coin flips, we have k=173 heads, a proportion of 0.865. By chance, this happens to be higher than the underlying true parameter value. After 200 flips, our posterior mean and its corresponding 95% interval are 0.848 (0.796, 0.893), which shows how strongly the likelihood has come to dominate the prior.
Markov Chain Monte Carlo (MCMC)
Attentive readers may have noticed that one buzzword frequently used in the context of applied Bayesian statistics – Markov Chain Monte Carlo (MCMC), an umbrella term for algorithms used for sampling from a posterior distribution – has been entirely absent from the coin flip example. Instead of using such MCMC algorithms, we have relied on an analytical solution for the posterior, exploiting the conjugacy of the beta and binomial distributions and the fact that this simple example with a single parameter (i.e., a unidimensional parameter space) allowed us to get the desired results with some quick and simple math.
For complex multi-dimensional posterior distributions, however, finding analytical solutions through integration becomes cumbersome, if not outright impossible. That’s where numerical approximation through MCMC algorithms comes in. MCMC algorithms are iterative computational processes that allow us to explore and describe posterior distributions. Essentially, we let Markov Chains wander through the parameter space. These should eventually, following an initial warmup period, converge to high-density regions in the underlying posterior distribution. The frequency of “times” (iterations) that the chain visits a given region of multidimensional parameter space gives a stochastic simulation of the posterior probability density in that region. Marginalizing the joint multidimensional posterior distribution with respect to a given single parameter, then, gives the posterior distribution for that parameter.
Some of the most frequently used MCMC algorithms include
- Gibbs Sampler: Draws iteratively through a complete set of conditional probability statements of each estimated parameter.
- Metropolis-Hastings: Considers a single multidimensional move on each iteration depending on the quality of the proposed candidate draw.
- Hamiltonian Monte Carlo (HMC), used in Stan:
The Hamiltonian Monte Carlo algorithm starts at a specified initial set of parameters \(\theta\); in Stan, this value is either user-specified or generated randomly. Then, for a given number of iterations, a new momentum vector is sampled and the current value of the parameter \(\theta\) is updated using the leapfrog integrator with discretization time \(\epsilon\) and number of steps \(L\) according to the Hamiltonian dynamics. Then a Metropolis acceptance step is applied, and a decision is made whether to update to the new state \((\theta^{\ast},\rho{\ast})\) or keep the existing state.
Readers interested in MCMC algorithms may want to consult the referenced section in the Stan Reference Manual and Chapter 9 of Gill (2015).
Applied Bayesian Statistics Using Stan and R
The Bayesian Workflow
Before we jump into the applications, we need to discuss the Bayesian workflow. Following Savage (2016), the typical workflow can be summarized as follows:
- Specification: Specify the full probability model including
- Data
- Parameters
- Priors
- Likelihood
- Model Building: Translate the model into code
- Validation: Validate the model with fake data
- Fitting: Fit the model to actual data
- Diagnosis: Check generic and algorithm-specific diagnostics to assess convergence
We thus start with the notational specification of a probability model and its translation into Stan code (steps 1 + 2). Unless we use ‘canned’ solutions, i.e. packages that generate accurate model code for us, validation of our Stan program using artifical data is utterly important: It allows us to test whether our model accurately retrieves prespecified parameters that we used for simulating the artificial data in the first place (step 3). Once this hurdle has been cleared, can we move on to fitting: The process of estimating the model parameters based on an actual data sample (step 4). In order to trust the corresponding estimates, we must then diagnose our estimates, using both generic and algorithm-specific diagnostic tools (step 5). In the following, we work through these steps using the example of the linear model.
Step 1: Specification
Stan requires that we be explicit about the known and unknown quantities in our model. This not only requires distinguishing the quantities we know from those we don’t know but also declaring object types (such as integer or real scalars, vectors, matrices, and arrays) and their respective dimensions. In this regard, it is worth quickly reviewing four different (yet fully equivalent) ways of the denoting the linear model formula: The scalar, row-vector, column-vector, and matrix forms.
- Scalar form: \[y_i = \beta_1 x_{i1} + \beta_2 x_{i2} + \beta_3 x_{i3} + \epsilon_i \text{ for all } i=1,...,N\]
- Row-vector form: \[y_i = \mathbf{x_i^{\prime}} \mathbf{\beta} + \epsilon_i \text{ for all } i=1,...,N\]
- Column-vector form: \[\mathbf{y} = \beta_1 \mathbf{x_{1}} + \beta_2 \mathbf{x_{2}} + \beta_3 \mathbf{x_{3}} + \mathbf{\epsilon}\]
- Matrix form: \[\mathbf{y = X \beta + \epsilon}\]
Of course, all four notations are based on the same quantities, which are compactly denoted by an outcome vector \(\mathbf{y}\), a model matrix \(\mathbf{X}\), a coefficient vector \(\beta\), and a vector of idiosyncratic error terms \(\epsilon\) in the matrix form. The first three variants only differ from the fourth in that they separate the multiplicative and additive operations entailed in \(\mathbf{X \beta}\) across the columns, rows, or cells of \(\mathbf{X}\). Hence, we can most compactly denote our objects according to the matrix form. Note that the model matrix typically contains a leading column of 1’s to multiply the intercept, \(\beta_1\). Therefore, \(x_{i1}\) in the scalar form and \(\mathbf{x_{1}}\) in the column-vector form are merely placeholders for 1’s and may be omitted notationally.
Let’s start by denoting the likelihood component of our model, summarized by the three core components of every generalized linear model:
- family: \(\mathbf{y} \sim \text{Normal}(\mu, \sigma)\)
- (inverse) link function: \(\text{id}(\mu) = \mu\)
- linear component: \(\mu = \mathbf{X} \beta\)
In words, we stipulate that the data be normally distributed with mean \(\mu = \mathbf{X} \beta\) and variance \(\sigma^2\). So what are our known and unknown quantities? The unknown quantities are
- \(\beta\), the coefficient vector
- \(\sigma\), the scale parameter of the normal
- \(\mu\), the location parameter of the normal
whereas the known quantities include
- \(\mathbf{y}\), the outcome vector
- \(\mathbf{X}\), the model matrix
as well as the dimensions of \(\mathbf{y}_{N \times 1}\) and \(\mathbf{X}_{N \times K}\) and the dimensions of \(\beta_{K \times 1}\), \(\sigma\) (a scalar), and \(\mu_{N \times 1}\).
Step 2: Model Building
Stan Program Blocks
Stan programs are defined in terms of several program blocks:
- Functions: Declare user-written functions
- Data: Declare all known quantities
- Transformed Data: Transform declared data inputs
- Parameters: Declare all unknown quantities
- Transformed Parameters: Transform declared parameters
- Model: Transform parameters, specify prior distributions and likelihoods to define posterior
- Generated Quantities: Generate quantities derived from the updated parameters without feedback into the likelihood
The parameter and model blocks are strictly required in order to define the sampling space and draw from the corresponding posterior distribution. Usually, applications also feature data inputs in the data block. The remaining four blocks are optional: Users may or may not specify their own functions, transform the initial data inputs, specify transformed parameters, or compute generated quantities.
Importantly, the blocks have different logics with respect to variable scopes. Functions, (transformed) data and (transformed) parameters have global scope: Once defined, they can be accessed and used in other program blocks. The scope of variables defined in the model and generated quantities blocks, in contrast, is local: Data and parameters defined here can only be accessed within the respective block.
The blocks also differ with respect to execution timing. Functions and (transformed) data are declared once and passed to each of the chains. The processing of the initially declared parameters in the transformed parameters block and definition of the posterior in the model block are evaluated at every so-called leapfrog step during every iteration of the HMC algorithm. The generated quantities block, in contrast, is only evaluated once per iteration, i.e., it uses the parameter values found as a result of the multi-step operations of the algorithm within each given iteration. Thus, for reasons of computational efficiency, quantities of interest that are derived from our (transformed) parameters but do not need to feed back into the likelihood (e.g., expected values or average marginal effects) should be declared and computed in the generated quantities block.
Model Scripts
For the following example, start with a blank script in your preferred code editor and save it as lm.stan. Using the stan suffix will enable syntax highlighting, formatting, and checking in RStudio and Emacs. Throughout the remainder of this subsection, we are coding in the Stan language: We separate declarations and statements by ;, type comments after //, and end each script with a blank line. The language is fully documented in the Stan Language Reference Manual and the Stan Language Functions Reference.
In the data block, we declare all known quantities, including data types, dimensions, and constraints:
- \(\mathbf{y}\), the outcome vector of length \(N\)
- \(\mathbf{X}\), the model matrix of dimensions \(N \times K\) (including a leading columns of 1’s to multiply the intercept)
data {
int<lower=1> N; // num. observations
int<lower=1> K; // num. predictors
matrix[N,K] x; // model matrix
vector[N] y; // outcome vector
}As we can see, we need to declare the integers \(N\) and \(K\) before we can specify the dimensions of the objects \(\mathbf{X}\) and \(\mathbf{y}\).
Next, we use the parameters block to declare all primitive unknown quantities, including their respective storage types, dimensions, and constraints:
- \(\beta\), the coefficient vector of length \(K\)
- \(\sigma\), the scale parameter of the normal, a non-negative real number
parameters {
vector[K] beta; // coef vector
real<lower=0> sigma; // scale parameter
}In the transformed parameters block, we declare and specify unknown transformed quantities, including storage types, dimensions, and constraints. In the following example, we use the transformed parameters block to compute our linear prediction, \(\mu = \mathbf{X} \beta\).
Note that we could just as well declare \(\mu\) in the model block – or not declare \(\mu\) as a variable at all but simply supply \(\mathbf{X} \beta\) in the log-likelihood instead. While all of these approaches would yield the same posterior, specifying \(\mu\) in the transformed parameters block makes an important difference: It ensures that \(\mu\) is stored as a global variable. As a result, samples of \(\mu\) will also be stored in the resulting output object.
transformed parameters {
vector[N] mu; // declare
mu = x * beta; // assign
}Lastly, we use the model block to declare and specify our sampling statements:
- \(\beta_k \sim \text{Normal}(0, 10) \text{ for } k = 1,...,K\); i.e., we assign every \(\beta\) coefficient an independent normal prior with a mean of 0 and standard deviation of 10
- \(\sigma \sim \text{Cauchy}^{+}(0, 5)\); i.e., we assign the scale parameter a Cauchy prior with a location of 0 and a scale of 5. Given that we have constrained the support for \(\sigma\) to non-negative values, the values will effectively be sampled from a half-Cauchy distribution
- \(\mathbf{y} \sim \text{Normal}(\mu, \sigma)\); i.e., we specify a normal log-likelihood, where every observation \(y_i\) follows a normal distribution with mean \(\mu_i\) and standard deviation \(\sigma\).
model {
// priors
beta ~ normal(0, 10); // priors for beta
sigma ~ cauchy(0, 5); // prior for sigma
// log-likelihood
target += normal_lpdf(y | mu, sigma);
}Putting all model blocks together in a single script then gives us our first Stan program.
Code: Full Stan program for the linear model
data {
int<lower=1> N; // num. observations
int<lower=1> K; // num. predictors
matrix[N,K] x; // model matrix
vector[N] y; // outcome vector
}
parameters {
vector[K] beta; // coef vector
real<lower=0> sigma; // scale parameter
}
transformed parameters {
vector[N] mu; // declare lin. pred.
mu = x * beta; // assign lin. pred.
}
model {
// priors
beta ~ normal(0, 10); // priors for beta
sigma ~ cauchy(0, 5); // prior for sigma
// log-likelihood
target += normal_lpdf(y | mu, sigma);
}
Before we proceed with the next steps of validation, fitting, and diagnosis, we discuss two extensions below: Specifying a weighted log-likelihood and standardizing our data as part of our program. These also illustrate the use of the functions, transformed data, and generated quantities blocks. Readers who wish to skip these extensions may continue reading here.
Extension 1: Weights
Using sampling, design, or poststratification weights is common practice among survey researchers. Weights ensure that observations do not contribute to the log-likelihood equally, but proportionally to their idiosyncratic weights. We can easily incorporate this feature into our Stan program for the linear model. The hack is fairly straightforward: It requires the definition of a new function as well as a single-line modification in the model block.
Before we define a function that allows us to retrieve the weighted log-likelihood, we first need to understand the built-in function for the unweighted log-likelihood, normal_lpdf, actually does. normal_lpdf defines the log of the normal probability density function (pdf) and sums across the resulting values of all observations, which returns a scalar:
\[ \mathtt{normal\_lpdf(y | mu, sigma)} = \sum_{i=1}^{N}\frac{1}{2} \log (2 \pi \sigma^2) \Big( \frac{y_i-\mu_i}{\sigma}\Big)\]
The fact that normal_lpdf sums across all observations is somewhat problematic for our purposes. To include weights, we need to weight every single entry in the log normal pdf prior to aggregation. In other words, we need a length-\(N\) vector of \(\mathtt{normal\_lpdf}\) values that we can then multiply with a length-\(N\) vector of weights before we sum across all observations. We therefore define a new function that returns the point-wise log normal pdf:
functions {
vector pw_norm(vector y, vector mu, real sigma) {
return -0.5 * (log(2 * pi() * square(sigma)) +
square((y - mu) / sigma));
}
}The remaining modifications are straightforward. First, we declare a vector of length \(N\) with idiosyncratic weights in the data block. We then use this vector in the model block, where we take the dot product of the weights and the vector of log-likelihood entries generated by pw_norm:
data {
...
vector<lower=0>[N] weights; // weights
}
...
model {
...
// weighted log-likelihood
target += dot_product(weights, pw_norm(y, mu, sigma));
}
The dot product returns the sum of the pairwise products of entries of both vectors, which gives us our weighted log-likelihood:
\[\mathtt{dot\_product(weights, pw\_norm(y, mu, sigma))} = \\ \sum_{i=1}^{N}\mathtt{weights}_i \times \frac{1}{2} \log (2 \pi \sigma^2) \Big( \frac{y_i-\mu_i}{\sigma}\Big)\]
Extension 2: Standardized Data
For our second extension, we follow this example from the Stan User’s Guide. Here, we use the transformed data block to standardize our outcome variable and predictors. Standardization can help us boost the efficiency of our model by allowing the model to converge toward the posterior distribution faster.
Our initial data inputs remain the same as in the original example: We declare \(N\), \(K\), \(\mathbf{X}\) and \(\mathbf{y}\) in the data block. We then use the transformed data block to standardize both \(\mathbf{y}\) and every column of \(\mathbf{X}\) (except the leading column of 1’s that multiplies the intercept). This means that for each variable, we first subtract its mean from each value and then divide the centered values by the variable’s standard deviation.
transformed data {
vector[K-1] sd_x; // std. dev. of predictors (excl. intercept)
vector[K-1] mean_x; // mean of predictors (excl. intercept)
vector[N] y_std; // std. outcome
matrix[N,K] x_std = x; // std. predictors
y_std = (y - mean(y)) / sd(y);
for (k in 2:K) {
mean_x[k-1] = mean(x[,k]);
sd_x[k-1] = sd(x[,k]);
x_std[,k] = (x[,k] - mean_x[k-1]) / sd_x[k-1];
}
}As a result of standardization, the posterior distributions of our estimated parameters change. This is, however, no cause for concern: We can simply use the generated quantities block to transform these parameters back to their original scale. Before we get there, we simply declare the parameters that we retrieve using the standardized data in the parameters block (named beta_std and sigma_std here) and complete the transformed parameters and model blocks analogous to the original example, using the standardized data, x_std and y_std, and the alternative parameters, beta_std, sigma_std and mu_std.
parameters {
vector[K] beta_std; // coef vector
real<lower=0> sigma_std; // scale parameter
}
transformed parameters {
vector[N] mu_std; // declare lin. pred.
mu_std = x_std * beta_std; // assign lin. pred.
}
model {
// priors
beta_std ~ normal(0, 10); // priors for beta
sigma_std ~ cauchy(0, 5); // prior for sigma
// log-likelihood
target += normal_lpdf(y_std | mu_std, sigma_std); // likelihood
}Using our draws for the alternative parameters beta_std and sigma_std, we can then use a little algebra to retrieve the original parameters:
- \(\beta_1 = \text{sd}(y) \Big(\beta_1^{\text{std}} - \sum_{k=2}^{K} \beta_k^{\text{std}} x_k^{\text{std}}\Big) + \bar{\mathbf{y}}\)
- \(\beta_k = \beta_k^{\text{std}} \frac{\text{sd}(y)}{\text{sd}(x_k)} \text{ for } k = 2,...,K\)
- \(\sigma = \text{sd}(y) \sigma^{\text{std}}\)
These calculations are implemented in the generated quantities block below:
generated quantities {
vector[K] beta; // coef vector
real<lower=0> sigma; // scale parameter
beta[1] = sd(y) * (beta_std[1] -
dot_product(beta_std[2:K], mean_x ./ sd_x)) + mean(y);
beta[2:K] = beta_std[2:K] ./ sd_x * sd(y);
sigma = sd(y) * sigma_std;
}Step 3: Validation
Validation and inference are two sides of the same coin. Whereas validation means that we estimate a Stan program on the basis of fake data, inference means that we estimate the same model on actual data to retrieve posterior distributions that speak to substantively meaningful questions.
Why start with fake data? Generating fake data allows us to mimic the data generating process underlying our model: By generating artificial predictors and arbitrarily choosing the ‘true’ model parameters, we can simulate the data generating process stipulated by our model to compute values of our outcome variable that we should observe given the predictors, the model parameters, and the likelihood function.
In turn, running our model using the simulated outcomes and artificial predictors should return parameter estimates close to the ‘true’ model parameters from which the outcome variable was simulated. If this is not the case, we know something went wrong: Assuming that our data-generating program is correct, there must be a problem in our model program. We should then go back to the model program and make sure that all parts of the script, including functions, data, parameterization, transformations, and the likelihood, are correctly specified.
The R code below implements our data-generating program: It simulates fake data which we will use to validate our Stan program for the linear model. After setting a seed for reproducibility, we simulate a model matrix \(\mathbf{X}\) with \(N=10000\) rows and \(K=5\) columns. Next to a leading column of 1’s, this matrix has four predictors generated from independent standard normal distributions. Next, we generate the true model parameters from their respective prior distributions: The \(\beta\) vector and the scale coefficient \(\sigma\). We then retrieve the linear prediction, \(\mu = \mathbf{X} \beta\), and use \(\mu\) and \(\sigma\) to simulate \(\mathbf{y}\) according to the likelihood of our model, \(\mathbf{y} \sim \text{N}(\mu,\sigma)\).
Code: Fake data generation
set.seed(20190417)
N.sim <- 10000L ### num. observations
K.sim <- 5L ### num. predictors
x.sim <- cbind( ### model matrix
rep(1, N.sim),
matrix(rnorm(N.sim * (K.sim - 1)), N.sim, (K.sim - 1))
)
beta.sim <- rnorm(K.sim, 0, 10) ### coef. vector
sigma.sim <- abs(rcauchy(1, 0, 5)) ### scale parameter
mu.sim <- x.sim %*% beta.sim ### linear prediction
y.sim <- rnorm(N.sim, mu.sim, sigma.sim) ### simulated outcome
Now that we have our fake data, we can set up for the next step, model validation. First, we load the rstan package and adjust some options to improve performance. Secondly, we collect the data we want to pass to our Stan program in a list named standat.sim. It is important that the object names in this list match the data declared in the data block of our program. This means that all data declared in the model block must be included in the list, named exactly as in the model script, and must have matching object and storage types (e.g., matrices should be matrices, integers should be integers, etc). Lastly, we compile our linear model using the stan_model() function. In this step, Stan uses a C++ compiler that translates the Stan program to C++ code.
## Setup
library(rstan)
rstan_options(auto_write = TRUE) ### avoid recompilation of models
options(mc.cores = parallel::detectCores()) ### parallelize across all CPUs
Sys.setenv(LOCAL_CPPFLAGS = '-march=native') ### improve execution time
## Data (see data block) as list
standat.sim <- list(
N = N.sim,
K = K.sim,
x = x.sim,
y = y.sim
)
## C++ Compilation
lm.mod <- stan_model(file = "code/lm.stan")We are now ready to run our model. Using the sampling() command, we retrieve a total of 2000 samples from the posterior distribution of pars = c("beta", "sigma"). Specifically, we let chains = 4L run in parallel across cores = 4L for iter = 2000L, each using algorithm = "NUTS", the No U-Turn Sampler variant of the Hamiltonian Monte Carlo algorithm. We then discard the first warmup = 1000L samples of each chain and thin the remaining samples of each chain by a factor of thin = 2L. For an explanation of additional options, see ?rstan::sampling.
lm.sim <- sampling(lm.mod, ### compiled model
data = standat.sim, ### data input
algorithm = "NUTS", ### algorithm
control = list( ### control arguments
adapt_delta = .85
),
save_warmup = FALSE, ### discard warmup samples
sample_file = NULL, ### no sample file
diagnostic_file = NULL, ### no diagnostic file
pars = c("beta", "sigma"), ### select parameters
iter = 2000L, ### iter per chain
warmup = 1000L, ### warmup period
thin = 2L, ### thinning factor
chains = 4L, ### num. chains
cores = 4L, ### num. cores
seed = 20190417) ### seedThe sampling() command generates a fitted Stan model of class stanfit, which we have stored as an object named lm.sim. Next to the samples from the posterior distributions of all chains (stored under lm.sim@sim$samples), this object contains extensive information on the specification of our Stan model and the (default) inputs to the sampling() command. The full structure of the stanfit object can be inspected below. ?rstan::stanfit also presents functions and methods for retrieving and summarizing the desired information from a stanfit object.
Code: Structure of the
stanfit object
str(lm.sim)## Formal class 'stanfit' [package "rstan"] with 10 slots
## ..@ model_name: chr "lm"
## ..@ model_pars: chr [1:4] "beta" "sigma" "mu" "lp__"
## ..@ par_dims :List of 4
## .. ..$ beta : num 5
## .. ..$ sigma: num(0)
## .. ..$ mu : num 10000
## .. ..$ lp__ : num(0)
## ..@ mode : int 0
## ..@ sim :List of 12
## .. ..$ samples :List of 4
## .. .. ..$ :List of 7
## .. .. .. ..$ beta[1]: num [1:500] -1.15 -0.953 -1.059 -1.144 -1.092 ...
## .. .. .. ..$ beta[2]: num [1:500] 5.55 5.55 5.72 5.69 5.67 ...
## .. .. .. ..$ beta[3]: num [1:500] 17.5 17.5 17.4 17.4 17.4 ...
## .. .. .. ..$ beta[4]: num [1:500] -0.914 -0.897 -0.979 -1.044 -1.105 ...
## .. .. .. ..$ beta[5]: num [1:500] -8.81 -8.75 -9.05 -8.91 -8.92 ...
## .. .. .. ..$ sigma : num [1:500] 11.7 12 11.9 11.9 11.9 ...
## .. .. .. ..$ lp__ : num [1:500] -38892 -38894 -38893 -38893 -38892 ...
## .. .. .. ..- attr(*, "test_grad")= logi FALSE
## .. .. .. ..- attr(*, "args")=List of 16
## .. .. .. .. ..$ append_samples : logi FALSE
## .. .. .. .. ..$ chain_id : num 1
## .. .. .. .. ..$ control :List of 12
## .. .. .. .. .. ..$ adapt_delta : num 0.85
## .. .. .. .. .. ..$ adapt_engaged : logi TRUE
## .. .. .. .. .. ..$ adapt_gamma : num 0.05
## .. .. .. .. .. ..$ adapt_init_buffer: num 75
## .. .. .. .. .. ..$ adapt_kappa : num 0.75
## .. .. .. .. .. ..$ adapt_t0 : num 10
## .. .. .. .. .. ..$ adapt_term_buffer: num 50
## .. .. .. .. .. ..$ adapt_window : num 25
## .. .. .. .. .. ..$ max_treedepth : int 10
## .. .. .. .. .. ..$ metric : chr "diag_e"
## .. .. .. .. .. ..$ stepsize : num 1
## .. .. .. .. .. ..$ stepsize_jitter : num 0
## .. .. .. .. ..$ enable_random_init: logi TRUE
## .. .. .. .. ..$ init : chr "random"
## .. .. .. .. ..$ init_list : NULL
## .. .. .. .. ..$ init_radius : num 2
## .. .. .. .. ..$ iter : int 2000
## .. .. .. .. ..$ method : chr "sampling"
## .. .. .. .. ..$ random_seed : chr "20190417"
## .. .. .. .. ..$ refresh : int 200
## .. .. .. .. ..$ sampler_t : chr "NUTS(diag_e)"
## .. .. .. .. ..$ save_warmup : logi FALSE
## .. .. .. .. ..$ test_grad : logi FALSE
## .. .. .. .. ..$ thin : int 2
## .. .. .. .. ..$ warmup : int 1000
## .. .. .. ..- attr(*, "inits")= num [1:10006] -0.5082 0.2635 -0.3416 -0.2569 -0.0969 ...
## .. .. .. ..- attr(*, "mean_pars")= num [1:10006] -1.07 5.58 17.53 -1.01 -8.9 ...
## .. .. .. ..- attr(*, "mean_lp__")= num -38894
## .. .. .. ..- attr(*, "adaptation_info")= chr "# Adaptation terminated\n# Step size = 0.678845\n# Diagonal elements of inverse mass matrix:\n# 0.0129191, 0.01"| __truncated__
## .. .. .. ..- attr(*, "elapsed_time")= Named num [1:2] 6.45 5.53
## .. .. .. .. ..- attr(*, "names")= chr [1:2] "warmup" "sample"
## .. .. .. ..- attr(*, "sampler_params")=List of 6
## .. .. .. .. ..$ accept_stat__: num [1:500] 0.799 0.844 0.82 0.802 0.959 ...
## .. .. .. .. ..$ stepsize__ : num [1:500] 0.679 0.679 0.679 0.679 0.679 ...
## .. .. .. .. ..$ treedepth__ : num [1:500] 3 3 2 3 3 2 3 3 3 2 ...
## .. .. .. .. ..$ n_leapfrog__ : num [1:500] 7 7 7 7 7 3 7 7 7 3 ...
## .. .. .. .. ..$ divergent__ : num [1:500] 0 0 0 0 0 0 0 0 0 0 ...
## .. .. .. .. ..$ energy__ : num [1:500] 38896 38896 38899 38898 38896 ...
## .. .. .. ..- attr(*, "return_code")= int 0
## .. .. ..$ :List of 7
## .. .. .. ..$ beta[1]: num [1:500] -0.946 -0.908 -1.164 -1.224 -1.206 ...
## .. .. .. ..$ beta[2]: num [1:500] 5.66 5.76 5.64 5.45 5.61 ...
## .. .. .. ..$ beta[3]: num [1:500] 17.6 17.6 17.8 17.7 17.4 ...
## .. .. .. ..$ beta[4]: num [1:500] -1.094 -1.124 -1.104 -1.111 -0.972 ...
## .. .. .. ..$ beta[5]: num [1:500] -8.78 -8.92 -9.11 -8.85 -9 ...
## .. .. .. ..$ sigma : num [1:500] 12 11.8 12 11.9 11.9 ...
## .. .. .. ..$ lp__ : num [1:500] -38894 -38893 -38897 -38894 -38892 ...
## .. .. .. ..- attr(*, "test_grad")= logi FALSE
## .. .. .. ..- attr(*, "args")=List of 16
## .. .. .. .. ..$ append_samples : logi FALSE
## .. .. .. .. ..$ chain_id : num 2
## .. .. .. .. ..$ control :List of 12
## .. .. .. .. .. ..$ adapt_delta : num 0.85
## .. .. .. .. .. ..$ adapt_engaged : logi TRUE
## .. .. .. .. .. ..$ adapt_gamma : num 0.05
## .. .. .. .. .. ..$ adapt_init_buffer: num 75
## .. .. .. .. .. ..$ adapt_kappa : num 0.75
## .. .. .. .. .. ..$ adapt_t0 : num 10
## .. .. .. .. .. ..$ adapt_term_buffer: num 50
## .. .. .. .. .. ..$ adapt_window : num 25
## .. .. .. .. .. ..$ max_treedepth : int 10
## .. .. .. .. .. ..$ metric : chr "diag_e"
## .. .. .. .. .. ..$ stepsize : num 1
## .. .. .. .. .. ..$ stepsize_jitter : num 0
## .. .. .. .. ..$ enable_random_init: logi TRUE
## .. .. .. .. ..$ init : chr "random"
## .. .. .. .. ..$ init_list : NULL
## .. .. .. .. ..$ init_radius : num 2
## .. .. .. .. ..$ iter : int 2000
## .. .. .. .. ..$ method : chr "sampling"
## .. .. .. .. ..$ random_seed : chr "20190417"
## .. .. .. .. ..$ refresh : int 200
## .. .. .. .. ..$ sampler_t : chr "NUTS(diag_e)"
## .. .. .. .. ..$ save_warmup : logi FALSE
## .. .. .. .. ..$ test_grad : logi FALSE
## .. .. .. .. ..$ thin : int 2
## .. .. .. .. ..$ warmup : int 1000
## .. .. .. ..- attr(*, "inits")= num [1:10006] 1.822 -1.301 0.447 1.979 1.029 ...
## .. .. .. ..- attr(*, "mean_pars")= num [1:10006] -1.07 5.57 17.53 -1.01 -8.91 ...
## .. .. .. ..- attr(*, "mean_lp__")= num -38894
## .. .. .. ..- attr(*, "adaptation_info")= chr "# Adaptation terminated\n# Step size = 0.598511\n# Diagonal elements of inverse mass matrix:\n# 0.0145165, 0.01"| __truncated__
## .. .. .. ..- attr(*, "elapsed_time")= Named num [1:2] 5.77 5.77
## .. .. .. .. ..- attr(*, "names")= chr [1:2] "warmup" "sample"
## .. .. .. ..- attr(*, "sampler_params")=List of 6
## .. .. .. .. ..$ accept_stat__: num [1:500] 0.86 0.936 0.878 0.98 0.996 ...
## .. .. .. .. ..$ stepsize__ : num [1:500] 0.599 0.599 0.599 0.599 0.599 ...
## .. .. .. .. ..$ treedepth__ : num [1:500] 3 3 3 3 3 3 3 3 2 3 ...
## .. .. .. .. ..$ n_leapfrog__ : num [1:500] 7 7 7 7 7 7 7 7 3 7 ...
## .. .. .. .. ..$ divergent__ : num [1:500] 0 0 0 0 0 0 0 0 0 0 ...
## .. .. .. .. ..$ energy__ : num [1:500] 38897 38897 38900 38897 38895 ...
## .. .. .. ..- attr(*, "return_code")= int 0
## .. .. ..$ :List of 7
## .. .. .. ..$ beta[1]: num [1:500] -0.887 -0.944 -1.084 -1.181 -1.011 ...
## .. .. .. ..$ beta[2]: num [1:500] 5.6 5.51 5.56 5.55 5.49 ...
## .. .. .. ..$ beta[3]: num [1:500] 17.5 17.5 17.6 17.4 17.6 ...
## .. .. .. ..$ beta[4]: num [1:500] -1.058 -1.033 -0.991 -0.96 -0.919 ...
## .. .. .. ..$ beta[5]: num [1:500] -8.85 -8.99 -8.81 -9.1 -8.88 ...
## .. .. .. ..$ sigma : num [1:500] 11.8 11.8 11.9 11.7 11.9 ...
## .. .. .. ..$ lp__ : num [1:500] -38892 -38892 -38891 -38894 -38891 ...
## .. .. .. ..- attr(*, "test_grad")= logi FALSE
## .. .. .. ..- attr(*, "args")=List of 16
## .. .. .. .. ..$ append_samples : logi FALSE
## .. .. .. .. ..$ chain_id : num 3
## .. .. .. .. ..$ control :List of 12
## .. .. .. .. .. ..$ adapt_delta : num 0.85
## .. .. .. .. .. ..$ adapt_engaged : logi TRUE
## .. .. .. .. .. ..$ adapt_gamma : num 0.05
## .. .. .. .. .. ..$ adapt_init_buffer: num 75
## .. .. .. .. .. ..$ adapt_kappa : num 0.75
## .. .. .. .. .. ..$ adapt_t0 : num 10
## .. .. .. .. .. ..$ adapt_term_buffer: num 50
## .. .. .. .. .. ..$ adapt_window : num 25
## .. .. .. .. .. ..$ max_treedepth : int 10
## .. .. .. .. .. ..$ metric : chr "diag_e"
## .. .. .. .. .. ..$ stepsize : num 1
## .. .. .. .. .. ..$ stepsize_jitter : num 0
## .. .. .. .. ..$ enable_random_init: logi TRUE
## .. .. .. .. ..$ init : chr "random"
## .. .. .. .. ..$ init_list : NULL
## .. .. .. .. ..$ init_radius : num 2
## .. .. .. .. ..$ iter : int 2000
## .. .. .. .. ..$ method : chr "sampling"
## .. .. .. .. ..$ random_seed : chr "20190417"
## .. .. .. .. ..$ refresh : int 200
## .. .. .. .. ..$ sampler_t : chr "NUTS(diag_e)"
## .. .. .. .. ..$ save_warmup : logi FALSE
## .. .. .. .. ..$ test_grad : logi FALSE
## .. .. .. .. ..$ thin : int 2
## .. .. .. .. ..$ warmup : int 1000
## .. .. .. ..- attr(*, "inits")= num [1:10006] -0.563 1.732 -1.012 1.965 -1.216 ...
## .. .. .. ..- attr(*, "mean_pars")= num [1:10006] -1.07 5.56 17.54 -1.01 -8.92 ...
## .. .. .. ..- attr(*, "mean_lp__")= num -38893
## .. .. .. ..- attr(*, "adaptation_info")= chr "# Adaptation terminated\n# Step size = 0.673836\n# Diagonal elements of inverse mass matrix:\n# 0.0157556, 0.01"| __truncated__
## .. .. .. ..- attr(*, "elapsed_time")= Named num [1:2] 6.92 4.42
## .. .. .. .. ..- attr(*, "names")= chr [1:2] "warmup" "sample"
## .. .. .. ..- attr(*, "sampler_params")=List of 6
## .. .. .. .. ..$ accept_stat__: num [1:500] 0.967 0.784 0.85 0.869 0.995 ...
## .. .. .. .. ..$ stepsize__ : num [1:500] 0.674 0.674 0.674 0.674 0.674 ...
## .. .. .. .. ..$ treedepth__ : num [1:500] 3 3 3 3 3 3 2 3 3 3 ...
## .. .. .. .. ..$ n_leapfrog__ : num [1:500] 7 7 7 7 7 7 3 7 7 7 ...
## .. .. .. .. ..$ divergent__ : num [1:500] 0 0 0 0 0 0 0 0 0 0 ...
## .. .. .. .. ..$ energy__ : num [1:500] 38894 38896 38895 38896 38892 ...
## .. .. .. ..- attr(*, "return_code")= int 0
## .. .. ..$ :List of 7
## .. .. .. ..$ beta[1]: num [1:500] -0.866 -1.339 -1.297 -1.238 -1.016 ...
## .. .. .. ..$ beta[2]: num [1:500] 5.23 5.63 5.7 5.56 5.64 ...
## .. .. .. ..$ beta[3]: num [1:500] 17.5 17.7 17.4 17.8 17.7 ...
## .. .. .. ..$ beta[4]: num [1:500] -1 -1 -1.3 -1.08 -1.08 ...
## .. .. .. ..$ beta[5]: num [1:500] -8.82 -8.9 -8.76 -8.99 -8.82 ...
## .. .. .. ..$ sigma : num [1:500] 11.7 11.9 11.9 11.9 11.8 ...
## .. .. .. ..$ lp__ : num [1:500] -38897 -38894 -38898 -38894 -38893 ...
## .. .. .. ..- attr(*, "test_grad")= logi FALSE
## .. .. .. ..- attr(*, "args")=List of 16
## .. .. .. .. ..$ append_samples : logi FALSE
## .. .. .. .. ..$ chain_id : num 4
## .. .. .. .. ..$ control :List of 12
## .. .. .. .. .. ..$ adapt_delta : num 0.85
## .. .. .. .. .. ..$ adapt_engaged : logi TRUE
## .. .. .. .. .. ..$ adapt_gamma : num 0.05
## .. .. .. .. .. ..$ adapt_init_buffer: num 75
## .. .. .. .. .. ..$ adapt_kappa : num 0.75
## .. .. .. .. .. ..$ adapt_t0 : num 10
## .. .. .. .. .. ..$ adapt_term_buffer: num 50
## .. .. .. .. .. ..$ adapt_window : num 25
## .. .. .. .. .. ..$ max_treedepth : int 10
## .. .. .. .. .. ..$ metric : chr "diag_e"
## .. .. .. .. .. ..$ stepsize : num 1
## .. .. .. .. .. ..$ stepsize_jitter : num 0
## .. .. .. .. ..$ enable_random_init: logi TRUE
## .. .. .. .. ..$ init : chr "random"
## .. .. .. .. ..$ init_list : NULL
## .. .. .. .. ..$ init_radius : num 2
## .. .. .. .. ..$ iter : int 2000
## .. .. .. .. ..$ method : chr "sampling"
## .. .. .. .. ..$ random_seed : chr "20190417"
## .. .. .. .. ..$ refresh : int 200
## .. .. .. .. ..$ sampler_t : chr "NUTS(diag_e)"
## .. .. .. .. ..$ save_warmup : logi FALSE
## .. .. .. .. ..$ test_grad : logi FALSE
## .. .. .. .. ..$ thin : int 2
## .. .. .. .. ..$ warmup : int 1000
## .. .. .. ..- attr(*, "inits")= num [1:10006] 0.6569 1.5602 -0.0956 -0.5841 -0.6011 ...
## .. .. .. ..- attr(*, "mean_pars")= num [1:10006] -1.06 5.56 17.54 -1.01 -8.92 ...
## .. .. .. ..- attr(*, "mean_lp__")= num -38894
## .. .. .. ..- attr(*, "adaptation_info")= chr "# Adaptation terminated\n# Step size = 0.611781\n# Diagonal elements of inverse mass matrix:\n# 0.0148783, 0.01"| __truncated__
## .. .. .. ..- attr(*, "elapsed_time")= Named num [1:2] 9.33 3.28
## .. .. .. .. ..- attr(*, "names")= chr [1:2] "warmup" "sample"
## .. .. .. ..- attr(*, "sampler_params")=List of 6
## .. .. .. .. ..$ accept_stat__: num [1:500] 0.99 0.995 0.754 1 0.88 ...
## .. .. .. .. ..$ stepsize__ : num [1:500] 0.612 0.612 0.612 0.612 0.612 ...
## .. .. .. .. ..$ treedepth__ : num [1:500] 3 3 3 3 2 2 3 3 2 3 ...
## .. .. .. .. ..$ n_leapfrog__ : num [1:500] 7 7 7 7 7 7 7 7 3 7 ...
## .. .. .. .. ..$ divergent__ : num [1:500] 0 0 0 0 0 0 0 0 0 0 ...
## .. .. .. .. ..$ energy__ : num [1:500] 38899 38899 38900 38899 38898 ...
## .. .. .. ..- attr(*, "return_code")= int 0
## .. ..$ chains : int 4
## .. ..$ iter : int 2000
## .. ..$ thin : int 2
## .. ..$ warmup : int 1000
## .. ..$ n_save : num [1:4] 500 500 500 500
## .. ..$ warmup2 : int [1:4] 0 0 0 0
## .. ..$ permutation:List of 4
## .. .. ..$ : int [1:500] 164 31 283 233 388 51 98 306 468 328 ...
## .. .. ..$ : int [1:500] 175 474 361 87 172 74 413 150 6 111 ...
## .. .. ..$ : int [1:500] 378 237 220 182 196 409 485 78 414 405 ...
## .. .. ..$ : int [1:500] 177 453 82 86 436 365 300 483 415 135 ...
## .. ..$ pars_oi : chr [1:3] "beta" "sigma" "lp__"
## .. ..$ dims_oi :List of 3
## .. .. ..$ beta : num 5
## .. .. ..$ sigma: num(0)
## .. .. ..$ lp__ : num(0)
## .. ..$ fnames_oi : chr [1:7] "beta[1]" "beta[2]" "beta[3]" "beta[4]" ...
## .. ..$ n_flatnames: int 7
## ..@ inits :List of 4
## .. ..$ :List of 3
## .. .. ..$ beta : num [1:5(1d)] -0.5082 0.2635 -0.3416 -0.2569 -0.0969
## .. .. ..$ sigma: num 0.587
## .. .. ..$ mu : num [1:10000(1d)] -0.9569 0.5379 -0.9832 -0.0281 -0.5676 ...
## .. ..$ :List of 3
## .. .. ..$ beta : num [1:5(1d)] 1.822 -1.301 0.447 1.979 1.029
## .. .. ..$ sigma: num 5.6
## .. .. ..$ mu : num [1:10000(1d)] 6.32 -1.64 4.65 -1.37 3.48 ...
## .. ..$ :List of 3
## .. .. ..$ beta : num [1:5(1d)] -0.563 1.732 -1.012 1.965 -1.216
## .. .. ..$ sigma: num 0.177
## .. .. ..$ mu : num [1:10000(1d)] -0.3923 1.2545 3.5917 0.0717 0.9679 ...
## .. ..$ :List of 3
## .. .. ..$ beta : num [1:5(1d)] 0.6569 1.5602 -0.0956 -0.5841 -0.6011
## .. .. ..$ sigma: num 5.47
## .. .. ..$ mu : num [1:10000(1d)] -3.0095 1.6166 0.0462 2.83 0.3272 ...
## ..@ stan_args :List of 4
## .. ..$ :List of 10
## .. .. ..$ chain_id : int 1
## .. .. ..$ iter : int 2000
## .. .. ..$ thin : int 2
## .. .. ..$ seed : int 20190417
## .. .. ..$ warmup : num 1000
## .. .. ..$ init : chr "random"
## .. .. ..$ algorithm : chr "NUTS"
## .. .. ..$ save_warmup: logi FALSE
## .. .. ..$ method : chr "sampling"
## .. .. ..$ control :List of 1
## .. .. .. ..$ adapt_delta: num 0.85
## .. ..$ :List of 10
## .. .. ..$ chain_id : int 2
## .. .. ..$ iter : int 2000
## .. .. ..$ thin : int 2
## .. .. ..$ seed : int 20190417
## .. .. ..$ warmup : num 1000
## .. .. ..$ init : chr "random"
## .. .. ..$ algorithm : chr "NUTS"
## .. .. ..$ save_warmup: logi FALSE
## .. .. ..$ method : chr "sampling"
## .. .. ..$ control :List of 1
## .. .. .. ..$ adapt_delta: num 0.85
## .. ..$ :List of 10
## .. .. ..$ chain_id : int 3
## .. .. ..$ iter : int 2000
## .. .. ..$ thin : int 2
## .. .. ..$ seed : int 20190417
## .. .. ..$ warmup : num 1000
## .. .. ..$ init : chr "random"
## .. .. ..$ algorithm : chr "NUTS"
## .. .. ..$ save_warmup: logi FALSE
## .. .. ..$ method : chr "sampling"
## .. .. ..$ control :List of 1
## .. .. .. ..$ adapt_delta: num 0.85
## .. ..$ :List of 10
## .. .. ..$ chain_id : int 4
## .. .. ..$ iter : int 2000
## .. .. ..$ thin : int 2
## .. .. ..$ seed : int 20190417
## .. .. ..$ warmup : num 1000
## .. .. ..$ init : chr "random"
## .. .. ..$ algorithm : chr "NUTS"
## .. .. ..$ save_warmup: logi FALSE
## .. .. ..$ method : chr "sampling"
## .. .. ..$ control :List of 1
## .. .. .. ..$ adapt_delta: num 0.85
## ..@ stanmodel :Formal class 'stanmodel' [package "rstan"] with 5 slots
## .. .. ..@ model_name : chr "lm"
## .. .. ..@ model_code : chr "data {\n int<lower=1> N; // num. observations\n int<lower=1> K; // num. predictors\n matrix[N, K] x; // desi"| __truncated__
## .. .. .. ..- attr(*, "model_name2")= chr "lm"
## .. .. ..@ model_cpp :List of 2
## .. .. .. ..$ model_cppname: chr "model362854de58e0_lm"
## .. .. .. ..$ model_cppcode: chr "// Code generated by Stan version 2.19.1\n\n#include <stan/model/model_header.hpp>\n\nnamespace model362854de58"| __truncated__
## .. .. ..@ mk_cppmodule:function (object)
## .. .. ..@ dso :Formal class 'cxxdso' [package "rstan"] with 7 slots
## .. .. .. .. ..@ sig :List of 1
## .. .. .. .. .. ..$ file36287001298d: chr(0)
## .. .. .. .. ..@ dso_saved : logi TRUE
## .. .. .. .. ..@ dso_filename: chr "file36287001298d"
## .. .. .. .. ..@ modulename : chr "stan_fit4model362854de58e0_lm_mod"
## .. .. .. .. ..@ system : chr "x86_64, mingw32"
## .. .. .. .. ..@ cxxflags : chr "CXXFLAGS=-O3 -Wno-unused-variable -Wno-unused-function"
## .. .. .. .. ..@ .CXXDSOMISC :<environment: 0x000000002647e630>
## ..@ date : chr "Wed Jan 22 14:29:59 2020"
## ..@ .MISC :<environment: 0x00000000268f4988>
For our purposes, (summaries of) the posterior distributions of the model parameters \(\beta\) and \(\sigma\) are of primary importance. In particular, we want to know if our Stan program accurately retrieves the ‘true’ parameters that we used to generate our artificial outcome data. We thus compare the true parameters
true.pars <- c(beta.sim, sigma.sim)
names(true.pars) <- c(paste0("beta[", 1:5, "]"), "sigma")
round(true.pars, 2L)## beta[1] beta[2] beta[3] beta[4] beta[5] sigma
## -1.03 5.50 17.33 -1.00 -8.88 11.98with the parameter estimates from our model:
print(lm.sim, pars = c("beta", "sigma"))## Inference for Stan model: lm.
## 4 chains, each with iter=2000; warmup=1000; thin=2;
## post-warmup draws per chain=500, total post-warmup draws=2000.
##
## mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat
## beta[1] -1.06 0 0.12 -1.30 -1.15 -1.06 -0.98 -0.84 1735 1
## beta[2] 5.57 0 0.12 5.35 5.49 5.57 5.65 5.79 1666 1
## beta[3] 17.54 0 0.12 17.31 17.46 17.53 17.62 17.76 1486 1
## beta[4] -1.01 0 0.12 -1.23 -1.09 -1.01 -0.93 -0.78 1472 1
## beta[5] -8.91 0 0.12 -9.15 -9.00 -8.92 -8.83 -8.67 1725 1
## sigma 11.82 0 0.08 11.66 11.77 11.82 11.88 11.99 1656 1
##
## Samples were drawn using NUTS(diag_e) at Wed Jan 22 14:29:59 2020.
## For each parameter, n_eff is a crude measure of effective sample size,
## and Rhat is the potential scale reduction factor on split chains (at
## convergence, Rhat=1).Unlike typical output after likelihood inference (i.e., point estimates and standard errors), we receive detailed summaries of the posterior distribution of each parameter. Here, we can think of the mean of a given posterior distribution as the counterpart to the point estimate, the sd as the counterpart to the standard error, and the 2.5% and 97.5% posterior percentiles as counterparts to the boundaries of a 95% confidence interval.
Ideally, we would want to see that the true parameter values lie near the center of their respective posterior distributions. As we can see above, the posterior means of all model parameters are close to the true parameter values and display only minor deviations. The question, of course, is how much deviation should have us worried. Also, one could suspect that the deviation from a given validation run is untypically large, e.g. due to a circumstantial selection of extreme ‘true’ parameter values when simulating the data generating process. Cook, Gelman, and Rubin (2006) therefore recommend running many replications of validation simulations, drawing different true parameter values from the respective prior distributions to generate a series of different simulated outcome values. They also propose test statistics that allow researchers to systematically analyze the magnitude of deviations between true parameters and posterior distributions.
Step 4: Inference
Having successfully validated our model, we can now run it on actual data. Model fitting works exactly as in the validation example above, using the same compiled Stan program and the sampling() command. The key difference is the data we supply. Instead of using simulated data, we now want to use an actual data set.
For the sake of illustration, we use the replication data for Table 1, Model 4, of Bischof and Wagner (2019), made available through the American Journal of Political Science Dataverse. The original analysis uses Ordinary Least Squares estimation to gauge the effect of the assassination of the populist radical right politician Pim Fortuyn prior to the Dutch Parliamentary Election in 2002 on micro-level ideological polarization. For this purpose, it analyzes 1551 respondents interviewed in the pre-election wave of the 2002 Dutch Parliamentary Election Study. The outcome variable contains squared distances of respondents’ left-right self-placement to the pre-election median self-placement of all respondents. The main predictor is a binary indicator whether the interview was conducted before or after Fortuyn’s assassination. The original analysis reports point estimates (standard errors) of 1.644 (0.036) for the intercept and -0.112 (0.076) for the before-/after indicator.
We now want to use our Stan program for the linear model to replicate these results. We thus retrieve the data from the AJPS dataverse and load them into R. Following this, we drop all but the three variables of interest, subset the data to the pre-election wave of the survey, and drop all incomplete rows. Now, we can easily extract the required input data for our linear model program (\(N\), \(K\), \(\mathbf{X}\) and \(\mathbf{y}\)) using the following code:
Code: Retrieving and managing the data
## Retrieve and manage data
bw.ajps19 <-
read.table(
paste0(
"https://dataverse.harvard.edu/api/access/datafile/",
":persistentId?persistentId=doi:10.7910/DVN/DZ1NFG/LFX4A9"
),
header = TRUE,
stringsAsFactors = FALSE,
sep = "\t",
fill = TRUE
) %>%
select(wave, fortuyn, polarization) %>% ### select relevant variables
subset(wave == 1) %>% ### subset to pre-election wave
na.omit() ### drop incomplete rows
x <- model.matrix(~ fortuyn, data = bw.ajps19)
y <- bw.ajps19$polarization
N <- nrow(x)
K <- ncol(x)
The rest then repeats the same steps we followed for validating our model with fake data: We collect the data objects in a list and pass the new list object, together with the compiled model, to the sampling() command.
Code: Fitting the model
## data as list
standat <- list(
N = N,
K = K,
x = x,
y = y)
## inference
lm.inf <- sampling(lm.mod, ### compiled model
data = standat, ### data input
algorithm = "NUTS", ### algorithm
control = list( ### control arguments
adapt_delta = .85
),
save_warmup = FALSE, ### discard warmup samples
sample_file = NULL, ### no sample file
diagnostic_file = NULL, ### no diagnostic file
pars = c("beta", "sigma"), ### select parameters
iter = 2000L, ### iter per chain
warmup = 1000L, ### warmup period
thin = 2L, ### thinning factor
chains = 4L, ### num. chains
cores = 4L, ### num. cores
seed = 20190417) ### seed
The resulting model output is summarized below:
print(lm.inf,
pars = c("beta", "sigma"),
digits_summary = 3L)## Inference for Stan model: lm.
## 4 chains, each with iter=2000; warmup=1000; thin=2;
## post-warmup draws per chain=500, total post-warmup draws=2000.
##
## mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat
## beta[1] 1.644 0.001 0.035 1.575 1.620 1.644 1.669 1.712 1763 1.000
## beta[2] -0.112 0.002 0.074 -0.250 -0.163 -0.113 -0.062 0.038 1898 0.999
## sigma 1.239 0.001 0.023 1.195 1.223 1.239 1.254 1.288 1891 1.000
##
## Samples were drawn using NUTS(diag_e) at Wed Jan 22 14:31:33 2020.
## For each parameter, n_eff is a crude measure of effective sample size,
## and Rhat is the potential scale reduction factor on split chains (at
## convergence, Rhat=1).As we can see, our posterior distribution summary closely matches the output reported in the original analysis. Analogously to the results reported in the article, which concludes that there is no significant positive effect of Fortuyn’s assassination on ideological polarization at the 95% level, we can also conclude that the probability of a positive effect for beta[2] is very low: Merely 6.5% of the corresponding posterior draws are greater than zero.
## Extract posterior samples for beta[2]
beta2_posterior <- extract(lm.inf)$beta[, 2]
## Probability that beta[2] is greater than zero
mean(beta2_posterior > 0)## [1] 0.065Step 5: Convergence Diagnostics
When using Markov Chain Monte Carlo (MCMC) algorithms for Bayesian Inference, we must always make sure that our samples from the parameter space are indicative of what we want – draws from the actual posterior distribution of interest. While simple, well-known models typically converge to the target posterior distribution fast, things may be more complicated for complex model programs. Multiple chains may not converge to the same posterior distribution; the exploration of the parameter space may be slow such that convergence is only achieved after a long warmup period; sequences of samples from the same chain may be highly correlated, necessitating long post-warmup runs for retrieving a reasonable large number of effective samples from the posterior distribution. These are just a few examples of things that can go wrong during Bayesian inference using MCMC methods. In the following, we introduce a number of diagnostic tools that allow users to assess the convergence of their Stan models.
Generic Diagnostics: Rhat and n_eff
By default, RStan reports two generic diagnostics when printing the summary output from a stanfit object (as in our linear model summaries above). The first of these is \(\hat{R}\), also known as the potential scale reduction statistic or as the Gelman-Rubin convergence diagnostic:
\[\small \widehat{Var}(\theta) = (1 - \frac{1}{\mathtt{n_{iter}}}) \underbrace{\Bigg(\frac{1}{ \mathtt{n_{chains}} (\mathtt{n_{iter}} - 1)} \sum_{j=1}^{\mathtt{n_{chains}}} \sum_{i=1}^{\mathtt{n_{iter}}} (\theta_{ij} - \bar{\theta_j})^2 \Bigg)}_{\text{Within chain var}} + \\ \frac{1}{\mathtt{n_{iter}}} \underbrace{\Bigg(\frac{\mathtt{n_{iter}}}{\mathtt{n_{chains} - 1}} \sum_{j=1}^{\mathtt{n_{chains}}} (\bar{\theta_j} - \bar{\bar{\theta}})^2\Bigg)}_{\text{Between chain var}}\]
This statistic combines information on the variation within and between chains, thus assessing whether each chain converged to a stationary target distribution and whether all chains converged to the same target distributions at the same time. Low values indicate that chains are stationary and mix well; high values are a cause for concern. As a rule of thumb, we want \(\hat{R}<1.1\) for all model parameters.
The second generic diagnostic tool is \(\mathtt{n_{eff}}\), the effective size of our posterior samples. A small effective sample size indicates high autocorrelation within chains, which in turn indicates that chains explore the posterior density very slowly and inefficiently. As a rule of thumb, we want the ratio of effective samples to total iterations to be \(\frac{\mathtt{n_{eff}}}{\mathtt{n_{iter}}} > 0.001\), i.e., we want a minimum of one effective sample per 1000 post-warmup iterations of our chains. However, such a rate is far from ideal because it requires that we run our sampler for many iterations to retrieve a sufficiently large effective sample size for valid inferences. In practice, we would thus likely explore the possibilities for efficiency tuning for improving the rate at which we retrieve effective samples.
Additional empirical diagnostics (see Gill 2015, Ch. 14.3.3) include
- Geweke Time-Series Diagnostic: Compare non-overlapping post-warmup portions of each chain to test within-convergence
- Heidelberger and Welch Diagnostic: Compare early post-warmup portion of each chain with late portion to test within-convergence
- Raftery and Lewis Integrated Diagnostic: Evaluates the full chain of a pilot run (requires that
save_warmup = TRUE) to estimate minimum required length of warmup and sampling
and are implemented as part of the coda package (Output Analysis and Diagnostics for MCMC). These can be used on stanfit objects after storing the posterior simulations as mcmc.listobjects, as illustrated below.
Code: Further generic diagnostics
## Stanfit to mcmc.list
lm.mcmc <- As.mcmc.list(lm.inf,
pars = c("beta", "sigma", "lp__"))
## Diagnostics
geweke.diag(lm.mcmc, frac1 = .1, frac2 = .5) ### Geweke## [[1]]
##
## Fraction in 1st window = 0.1
## Fraction in 2nd window = 0.5
##
## beta[1] beta[2] sigma lp__
## 0.9361 -1.9100 1.1021 1.8148
##
##
## [[2]]
##
## Fraction in 1st window = 0.1
## Fraction in 2nd window = 0.5
##
## beta[1] beta[2] sigma lp__
## 0.7879 -0.9223 0.6536 0.9696
##
##
## [[3]]
##
## Fraction in 1st window = 0.1
## Fraction in 2nd window = 0.5
##
## beta[1] beta[2] sigma lp__
## -0.5272 -1.2241 0.0946 0.4136
##
##
## [[4]]
##
## Fraction in 1st window = 0.1
## Fraction in 2nd window = 0.5
##
## beta[1] beta[2] sigma lp__
## -0.9422 0.8782 -0.1088 -2.6450heidel.diag(lm.mcmc, pvalue = .1) ### Heidelberger-Welch## [[1]]
##
## Stationarity start p-value
## test iteration
## beta[1] passed 1 0.482
## beta[2] passed 1 0.594
## sigma passed 1 0.548
## lp__ passed 1 0.308
##
## Halfwidth Mean Halfwidth
## test
## beta[1] passed 1.643 0.00317
## beta[2] passed -0.111 0.00740
## sigma passed 1.240 0.00206
## lp__ passed -2532.284 0.10164
##
## [[2]]
##
## Stationarity start p-value
## test iteration
## beta[1] passed 1 0.627
## beta[2] passed 1 0.817
## sigma passed 1 0.883
## lp__ passed 1 0.688
##
## Halfwidth Mean Halfwidth
## test
## beta[1] passed 1.645 0.00290
## beta[2] passed -0.111 0.00612
## sigma passed 1.240 0.00212
## lp__ passed -2532.403 0.12362
##
## [[3]]
##
## Stationarity start p-value
## test iteration
## beta[1] passed 1 0.2911
## beta[2] passed 51 0.0653
## sigma passed 1 0.9587
## lp__ passed 1 0.6963
##
## Halfwidth Mean Halfwidth
## test
## beta[1] passed 1.64 0.00311
## beta[2] passed -0.11 0.00719
## sigma passed 1.24 0.00195
## lp__ passed -2532.39 0.13209
##
## [[4]]
##
## Stationarity start p-value
## test iteration
## beta[1] passed 1 0.593
## beta[2] passed 1 0.638
## sigma passed 1 0.526
## lp__ passed 1 0.118
##
## Halfwidth Mean Halfwidth
## test
## beta[1] passed 1.646 0.00310
## beta[2] passed -0.114 0.00665
## sigma passed 1.239 0.00195
## lp__ passed -2532.447 0.11456raftery.diag(lm.mcmc, ### Raftery-Lewis
q = 0.025,
r = 0.005,
s = 0.95,
converge.eps = 0.001)## [[1]]
##
## Quantile (q) = 0.025
## Accuracy (r) = +/- 0.005
## Probability (s) = 0.95
##
## You need a sample size of at least 3746 with these values of q, r and s
##
## [[2]]
##
## Quantile (q) = 0.025
## Accuracy (r) = +/- 0.005
## Probability (s) = 0.95
##
## You need a sample size of at least 3746 with these values of q, r and s
##
## [[3]]
##
## Quantile (q) = 0.025
## Accuracy (r) = +/- 0.005
## Probability (s) = 0.95
##
## You need a sample size of at least 3746 with these values of q, r and s
##
## [[4]]
##
## Quantile (q) = 0.025
## Accuracy (r) = +/- 0.005
## Probability (s) = 0.95
##
## You need a sample size of at least 3746 with these values of q, r and sAlgorithm-specific Diagnostics
Next to these generic empirical diagnostics, Stan also offers algorithm-specific diagnostic tools after Hamiltonian Monte Carlo sampling. In the words of the developers,
Hamiltonian Monte Carlo provides not only state-of-the-art sampling speed, it also provides state-of-the-art diagnostics. Unlike other algorithms, when Hamiltonian Monte Carlo fails it fails sufficiently spectacularly that we can easily identify the problems.
These diagnostic tools can identify various problems. The first of these is divergent transitions after warmup, which can be tackled by increasing adapt_delta (the target acceptance rate) or by optimizing the model program (see efficiency tuning). Tackling this problem is imperative because even single divergent transitions can undermine the validity of our estimates. Another problem is exceeding the maximum treedepth, which can be tackled by increasing max_treedepth. Unlike divergent transitions, exceeding the maximum treedepth is merely an efficiency concern that slows down computation time. Moreover, the check_hmc_diagnostics() command offers extensive diagnostics summary for stanfit objects. For further information, see the Guide to Stan’s warnings.
Visual Diagnostics
In addition to empirical diagnostics, users can also use visual diagnostics. One way of doing so is using shinystan. Maintained by the Stan Development Team, this package launches a local Shiny web application that allows users to interactively explore visual and numerical diagnostics for posterior draws and algorithm performance in their web browser. Among other things, this tool allows users to view and export multiple variants of visual diagnostics as well as uni- and multivariate graphical and numerical posterior summaries of parameters Its additional functionality includes the generate_quantity() function, which creates a new parameter as a function of one or two existing parameters, and the deploy_shinystan() function, which allows users to deploy a ‘ShinyStan’ app on shinyapps.io. To explore shinystan, users can access a demo using launch_shinystan_demo().
An alternative is bayesplot. This package offers a vast selection of visual diagnostics for stanfit objects, including various diagnostics for the No-U-Turn-Sampler (divergent transitions, energy, Bayesian fraction of missing information) and generic MCMC diangostics (\(\hat{R}\), \(\mathtt{n_{eff}}\), autocorrelation, and mixing through trace plots). For full functionality, examples, and vignettes, see these examples on GitHub, the CRAN Vignettes, and the available_mcmc() function.
The example below illustrates how bayesplot can be used to assess within and between chain convergence using traceplots. Traceplots show the post-warmup samples from all chains against as a time series (plotted against the chains’ index). In case of convergence, we should see that each chain and all chains fluctuate around the same value throughout the post-warmup period. For instance, no single chain should shift its center of gravity from low to high values over the course of the chain and all chains should mix around the same range of values. As we can see below, this is the case for all three model parameters.
## Extract posterior draws from stanfit object
lm.post.draws <- extract(lm.inf, permuted = FALSE)
## Traceplot
mcmc_trace(lm.post.draws, pars = c("beta[1]", "beta[2]", "sigma"))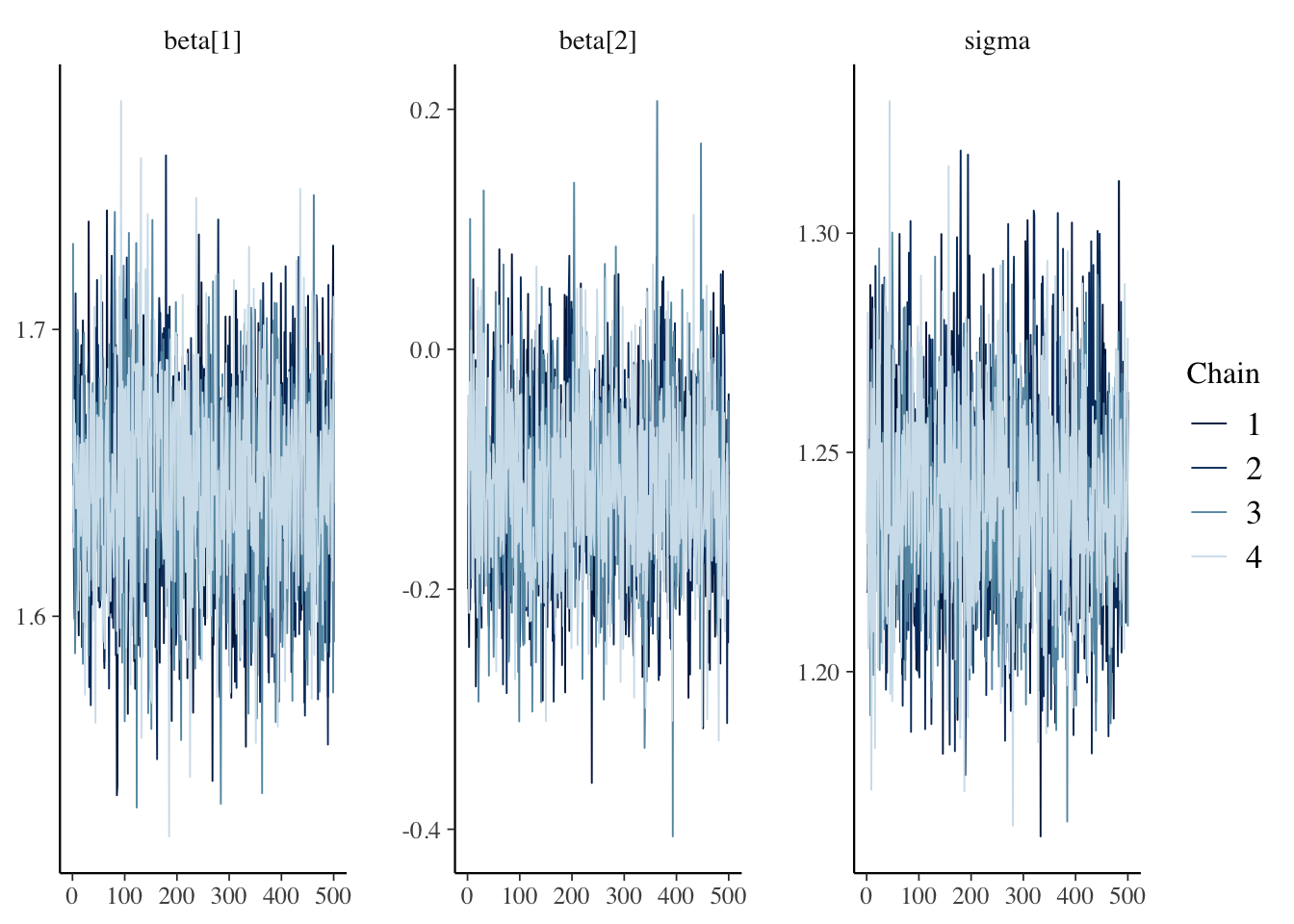
Additional Interfaces
One of the key advantages of Stan over other programming languages for Bayesian inference is the availability of additional interfaces that offer ‘canned solutions’, i.e., pre-programmed functions for the estimation and analysis of widely used statistical models. Two of the most popular interfaces are rstanarm and brms.
rstanarm
rstanarm has been developed by Stan Development Team members Jonah Gabry and Ben Goodrich, along with numerous contributors. In the words of its developers,
“rstanarm is an R package that emulates other R model-fitting functions but uses Stan (via the rstan package) for the back-end estimation. The primary target audience is people who would be open to Bayesian inference if using Bayesian software were easier but would use frequentist software otherwise.”
Source: https://mc-stan.org/rstanarm/
rstanarm covers a broad range of model types, including
stan_lm,stan_aov,stan_biglm: Regularized linear models; similar tolmandaovstan_glm,stan_glm.nb: Generalized linear models; similar toglmstan_glmer,stan_glmer.nb,stan_lmer: Generalized linear models with group-specific terms; similar toglmer,glmer.nb, andlmer(lme4)stan_nlmer: Nonlinear models with group-specific terms; similarnlmer(lme4)stan_gamm4: Generalized linear additive models with optional group-specific terms; similar togamm4(gamm4)stan_polr: Ordinal regression models; similar topolr(MASS)stan_betareg: Beta regression models; similar tobetareg(betareg)stan_clogit: Conditional logistics regression models; similar toclogit(survival)stan_mvmer: Multivariate generalized linear models with correlated group-specific terms; a multivariate form ofstan_glmerstan_jm: Shared parameter joint models for longitudinal and time-to-event models
The package comes with its own web site which features an extensive collection of vignettes and tutorials.
brms
Developed by Paul-Christian Bürkner, brms offers another interface for statistical modeling using the Stan back-end:
“The brms package provides an interface to fit Bayesian generalized (non-)linear multivariate multilevel models using Stan, which is a C++ package for performing full Bayesian inference (see http://mc-stan.org/). The formula syntax is very similar to that of the package lme4 to provide a familiar and simple interface for performing regression analyses.”
The package has extensive functionality, supporting a large collection of model types, including
- linear models
- robust linear models
- binomial models
- categorical models
- multinomial models
- count data models
- survival models
- ordinal models
- zero-inflated and hurdle models
- generalized additive models
- non-linear models
Source: BRMS Overview Vignette
In addition to a detailed overview and a web site that features extensive documentation, vignettes, and tutorials, brms comes with some additional useful features. These include
marginal_effects(): Display marginal effects of one or more numeric and/or categorical predictors including two-way interaction effectsbrm_multiple(): Inference across imputations generated bymiceprior to model fitting in brmsmi(): Fully Bayesian imputation during model fitting
Concluding Remarks
Reproducibility
Reproducibility is an important standard for the quality of quantitative research. Unfortunately, one and the same Stan program can produce (slightly) different parameter estimates even when identical seeds are supplied to the sampler. According to the Stan Reference Manual, Stan is only guaranteed to fully reproduce results if all of the following are held constant:
- Stan version
- Stan interface (RStan, PyStan, CmdStan) and version, plus version of interface language (R, Python, shell)
- versions of included libraries (Boost and Eigen)
- operating system version
- computer hardware including CPU, motherboard and memory
- C++ compiler, including version, compiler flags, and linked libraries
- same configuration of call to Stan, including random seed, chain ID, initialization and data
Source: Stan Reference Manual
While deviations from user’s original results due to version mismatches are likely to be minor, users are thus advised to document their software and hardware configurations.
Summary
Stan offers a powerful tool for statistical inference using not only full Bayesian inference but also variational inference or penalized maximum likelihood estimation. It provides an intuitive language for statistical modeling that accommodates most, though perhaps not every user’s needs (most notably, no discrete parameters). Stan greatly facilitates parallelization and convergence diagnosis. R packages like rstanarm and brms offer a vast array of ‘canned’ solutions. These come with the benefit of making Bayesian inference easily accessible to a broad audience but arguably come with pitfalls when users overly rely on default settings.
Applied modeling beyond canned solutions may require some specialized knowledge, e.g. when it comes to Jacobian adjustments when sampling transformed parameters, the marginalization of discrete parameters, or effiency tuning. Furthermore, Stan’s primary algorithm, the NUTS variant of HMC (see Hoffman and Gelman 2014), tends to remain a black box even for long-time Bayesian practitioners. This should, however, by no means discourage aspiring users from learning Stan. After all, one of the core strengths of the Stan project is its extensive documentation, its vast collection of freely available learning materials, and its active and responsive online community.
About the Presenter
Denis Cohen is a postdoctoral fellow in the Data and Methods Unit at the Mannheim Centre for European Social Research (MZES), University of Mannheim, and one of the organizers of the MZES Social Science Data Lab. His research focus lies at the intersection of political preference formation, electoral behavior, and political competition. His methodological interests include quantitative approaches to the analysis of clustered data, measurement models, data visualization, strategies for causal identification, and Bayesian statistics.
Further Reading
- Aczel, Balazs, Rink Hoekstra, Andrew Gelman, Eric-Jan Wagenmakers, Irene G. Klugkist, Jeffrey N. Rouder, Joachim Vandekerckhove, Michael D. Lee, Richard D. Morey, Wolf Vanpaemel, Zoltan Dienes and Don van Ravenzwaaij (2020). Discussion points for Bayesian inference. Nature Human Behaviour.
- Schlierholz, Malte. SSDL Input Talk Fundamentals in Bayesian Statistics. December 14, 2016.
- Stan Best Practices
- staudtlex.de – Alex Staudt’s web site, with R and C++ implementations of the Metropolis-Hastings algorithm
- Tutorial: Robust Statistical Workflow with RStan
References
Bischof, Daniel, and Markus Wagner. 2019. “Do Voters PolarizeWhen Radical Parties Enter Parliament.” American Journal of Political Science. https://onlinelibrary.wiley.com/doi/abs/10.1111/ajps.12449.
Bürkner, Paul-Christian. 2017a. “Advanced Bayesian Multilevel Modeling with the R Package brms.” The R Journal 10 (1): 395–411. http://arxiv.org/abs/1705.11123.
———. 2017b. “brms : An R Package for Bayesian Multilevel Models Using Stan.” Journal of Statistical Software 80 (1). https://doi.org/10.18637/jss.v080.i01.
Carpenter, Bob, Andrew Gelman, Matthew D Hoffman, Daniel Lee, Ben Goodrich, Michael Betancourt, Marcus Brubaker, Jiqiang Guo, Peter Li, and Allen Riddell. 2017. “Stan : A Probabilistic Programming Language.” Journal of Statistical Software 76 (1). https://doi.org/10.18637/jss.v076.i01.
Cook, Samantha R., Andrew Gelman, and Donald B. Rubin. 2006. “Validation of software for Bayesian models using posterior quantiles.” Journal of Computational and Graphical Statistics 15 (3): 675–92. https://doi.org/10.1198/106186006X136976.
Gill, Jeff. 2015. Bayesian Methods. A Social and Behavioral Sciences Approach. 3rd ed. Boca Raton, FL: CRC Press.
Gill, Jeff, and Christopher Witko. 2013. “Bayesian analytical methods: A methodological prescription for public administration.” Journal of Public Administration Research and Theory 23 (2): 457–94. https://doi.org/10.1093/jopart/mus091.
Hoffman, Matthew D., and Andrew Gelman. 2014. “The No-U-Turn Sampler: Adaptively setting path lengths in Hamiltonian Monte Carlo.” Journal of Machine Learning Research 15: 1593–1623. http://arxiv.org/abs/1111.4246.
Stan Development Team. 2019. “Stan Functions Reference. Version 2.19.”
